Welcome to the forums at seaphages.org. Please feel free to ask any questions related to the SEA-PHAGES program. Any logged-in user may post new topics and reply to existing topics. If you'd like to see a new forum created, please contact us using our form or email us at info@seaphages.org.
Recent Activity
All posts created by DanRussell
| Link to this post | posted 19 Nov, 2018 17:51 | |
|---|---|
|
|
Hi Bill, Since BabeRuth sounds like the outlier here, I went back and took a look at the BabeRuth sequencing data. Here's a pic of that region. 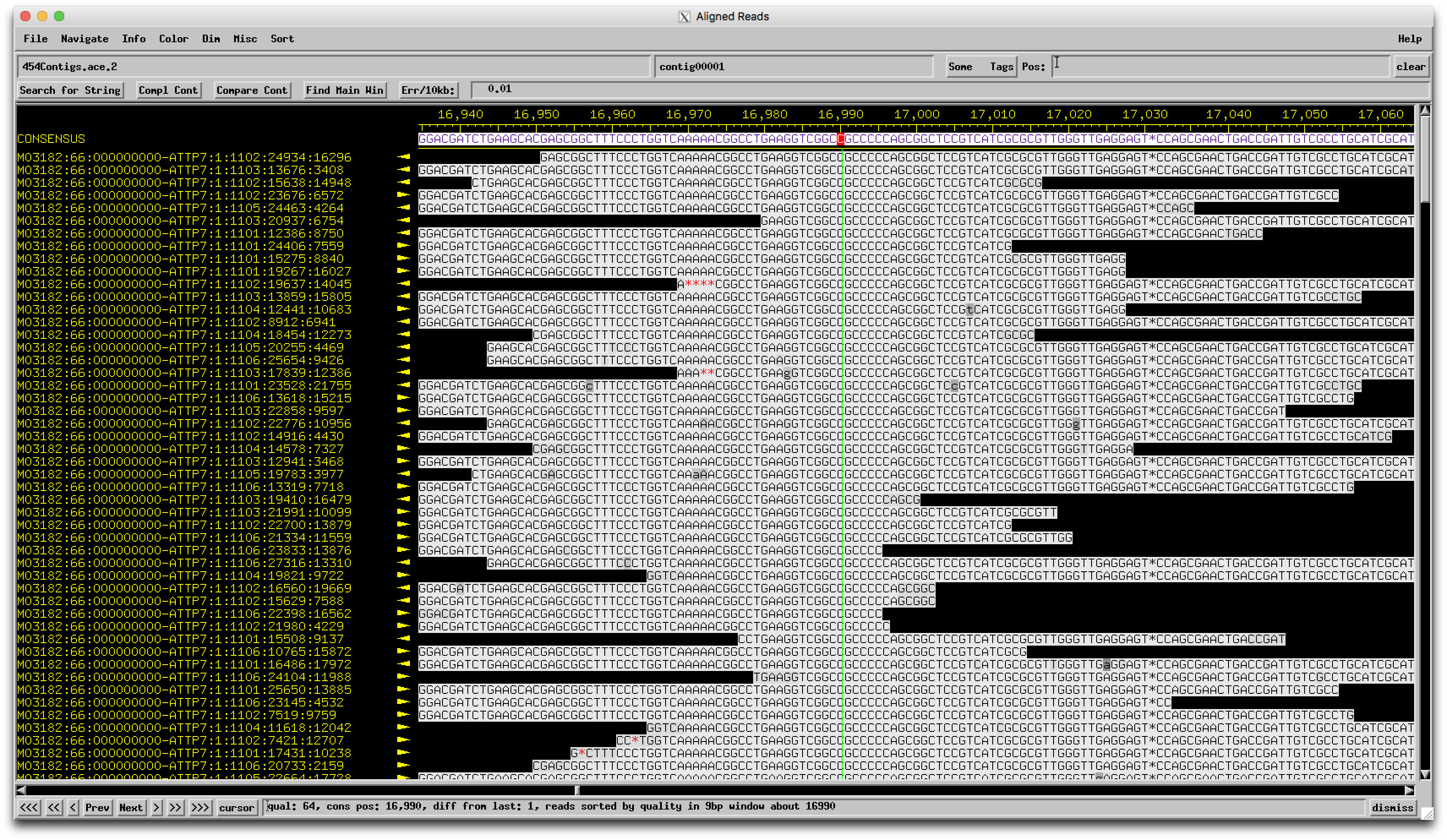 Very solid coverage and agreement that there are indeed 5 Cs here, not 4 like the others with similar genomes. This is a real biological feature, not a sequencing error! I can't say for sure of course whether this is really a frameshift (in which case the protein would still be made) or just a mutation that truncates the protein (more likely), but the underlying DNA sequence at least is correct. Good eye, and this is definitely the kind of thing that's worth checking. –Dan |
Posted in: Frameshifts and Introns → Putative Frameshift in a Reverse gene in N-Cluster Phage BabeRuth
| Link to this post | posted 26 Jun, 2018 20:18 | |
|---|---|
|
|
Hey Germán, I was able to find the original data for Anglerfish on one of our old machines. Turns out it is indeed a sequencing error. About 50% of the reads had 4 Cs instead of 3 Cs there, but the consensus was called with 3 Cs. Since it was sequenced in the early days of Ion Torrent, we weren't as good at picking up those errors, but after that definitely confirmed that in cases like that, the longer string was correct. I've added a fourth C to that position in the PhagesDB fasta file. I'll be in touch via email about how to move your existing annotation info to the new sequence. –Dan |
Posted in: Frameshifts and Introns → New frameshift in an A1 phage?
| Link to this post | posted 15 Jun, 2018 15:53 | |
|---|---|
|
|
Pollenz Ah, SSL error. Did you see this? https://seaphages.org/forums/topic/4436/?page=1#post-5805 –Dan |
Posted in: DNA Master → Important DNA Master Update
| Link to this post | posted 13 Jun, 2018 15:04 | |
|---|---|
|
|
Hi Rick, Did you update DNA Master? Try running an update. (Help–>Update DNA Master) Then restart and check your version. (Help–>About) The latest version is: Version 5.23.2 Build 2583 15 May 2018 –Dan |
Posted in: DNA Master → Important DNA Master Update
| Link to this post | posted 12 Jun, 2018 12:59 | |
|---|---|
|
|
Hey Germán, Since this one was sequenced a while ago, I think I should probably check the sequence in this region to see if it's a sequencing error. I'll get to that ASAP. –Dan |
Posted in: Frameshifts and Introns → New frameshift in an A1 phage?
| Link to this post | posted 04 Jun, 2018 21:12 | |
|---|---|
|
|
Hi Ann, I'm not sure there is. My recent experiences with WINE have varied, and almost always the size of the genome is a determining factor in how well WINE works. A 50 kb genome loads in a few seconds, but a 100 kb genome never works. Not helpful, obviously, but others may have better suggestions. You may be able to request a loaner laptop for the workshop. I'd ask Vic and/or Billy. –Dan |
Posted in: DNA Master → Annotating Cluster C1 phage onn Macbook Pro
| Link to this post | posted 26 Apr, 2018 13:55 | |
|---|---|
|
|
Hi Andrea, As Debbie said, the "master" copy of Phamerator gets updated about once a week. PhagesDB checks every night to see if a new version exists and if so, installs it. Online Phamerator does the same, but might not check every night, which is why things may appear out-of-sync for a day or two. But thanks for letting us know, and I'll double-check that everything is working as intended. –Dan |
Posted in: Web Phamerator → Pham Number Discrepancy
| Link to this post | posted 27 Feb, 2018 16:55 | |
|---|---|
|
|
FROM AN NIH LIST Dear All, An ORISE fellow position is available in my laboratory for a project involving virology and bioinformatics knowledge including computer language. This position provides a training opportunity for enthusiasts interested in developing bioinformatics tools and databases for virus detection, annotation, and characterization. The overall goal of the project is cell line and vaccine safety. Individuals that meet the requirements should submit their CV and contact information for three references. Thank you. Regards, Arifa Khan Supervisory Microbiologist Laboratory of Retrovirus Research Division of Viral Products Office of Vaccines Research and Review Center for Biologics Evaluation and Research U.S. Food and Drug Administration 10903 New Hampshire Ave Bldg. 52-72, Room 1216 Silver Spring, MD 20993 Tel no. 240-402-9631 Email: arifa.khan@fda.hhs.gov |
Posted in: General Message Board → ORISE Fellow Position at FDA
| Link to this post | posted 26 Jan, 2018 20:48 | |
|---|---|
|
|
GregFrederick@letu.edu Hi Greg, To be clear: your problem is with BLAST, not auto annotation itself? Meaning you get all the features, but only some of them have BLAST results? BLASTing all genes can sometimes end up with genes being skipped, especially if done during peak hours. What phage are you working on? –Dan |
Posted in: DNA Master → Auto-annotation fix for fall 2017 and later
| Link to this post | posted 26 Jan, 2018 20:46 | |
|---|---|
|
|
ptsourkas Hi Philippos, Is your DNA Master up to date? If you run Update does all look good? Any other weird errors popping up? I think the servers are working. You've entered the proper access code in the linked document? –Dan |
Posted in: DNA Master → Auto-annotation fix for fall 2017 and later
