Welcome to the forums at seaphages.org. Please feel free to ask any questions related to the SEA-PHAGES program. Any logged-in user may post new topics and reply to existing topics. If you'd like to see a new forum created, please contact us using our form or email us at info@seaphages.org.
Recent Activity
All posts created by DanRussell
| Link to this post | posted 20 Jan, 2026 15:54 | |
|---|---|
|
|
Hi all, Many folks ask each year about getting extra phage genomes sequenced beyond the two per section we sequence as part of SEA-PHAGES support. We at Pitt cannot do extra sequencing for payment, but there are a number of commercial options out there. That said, you'll want to try to avoid transposon-based library preps when getting new phage genomes sequenced, and one option that has experience with phages and will use the preferred library prep method is NC State. NC State has been doing phage sequencing for a number of years with over 230 of the sequences on PhagesDB having been sequenced there. The limitation they're working with, however, is getting up to a "critical mass" of phage samples in order to make this more niche type of run worthwhile. So turnaround can be slower if fewer people are sending samples. Which is to say: if enough of you are interested in having more of your DNA sequenced, and you reach out to NC State (by contacting Kelly Sides, kafridey@ncsu.edu), you may get good quality data back in a relatively short amount of time. Kelly can answer any questions you have about their services. Feel free to respond here if you're thinking of getting more samples sequenced to connect and get those numbers up. And remember that we at Pitt are always happy to help with the assembly and QC post-sequencing, even when things are sequenced elsewhere! –Dan |
| Link to this post | posted 13 Jan, 2026 18:58 | |
|---|---|
|
|
Hi Dave, I have an Apple M3 MacBook, and I use VMWare Fusion to successfully run DNA Master. –Dan |
Posted in: DNA Master → Emulator Choice
| Link to this post | posted 11 Mar, 2025 15:07 | |
|---|---|
|
|
Hi Kieran, Yes, for any phages we finished from March 1, 2024, onwards, here are the details for read trimming. Reads were trimmed and filtered by: cutadapt 4.7 (using the option: –nextseq-trim 30) and skewer 0.2.2 (using the options: -q 20 -Q 30 -n -l 50) For assembly, we're now using: Unicycler version 0.5.1 For QC, it's still: Consed version 29 I've also added some more details about sequencing to phage pages on PhagesDB. That might be helpful when writing MRAs. 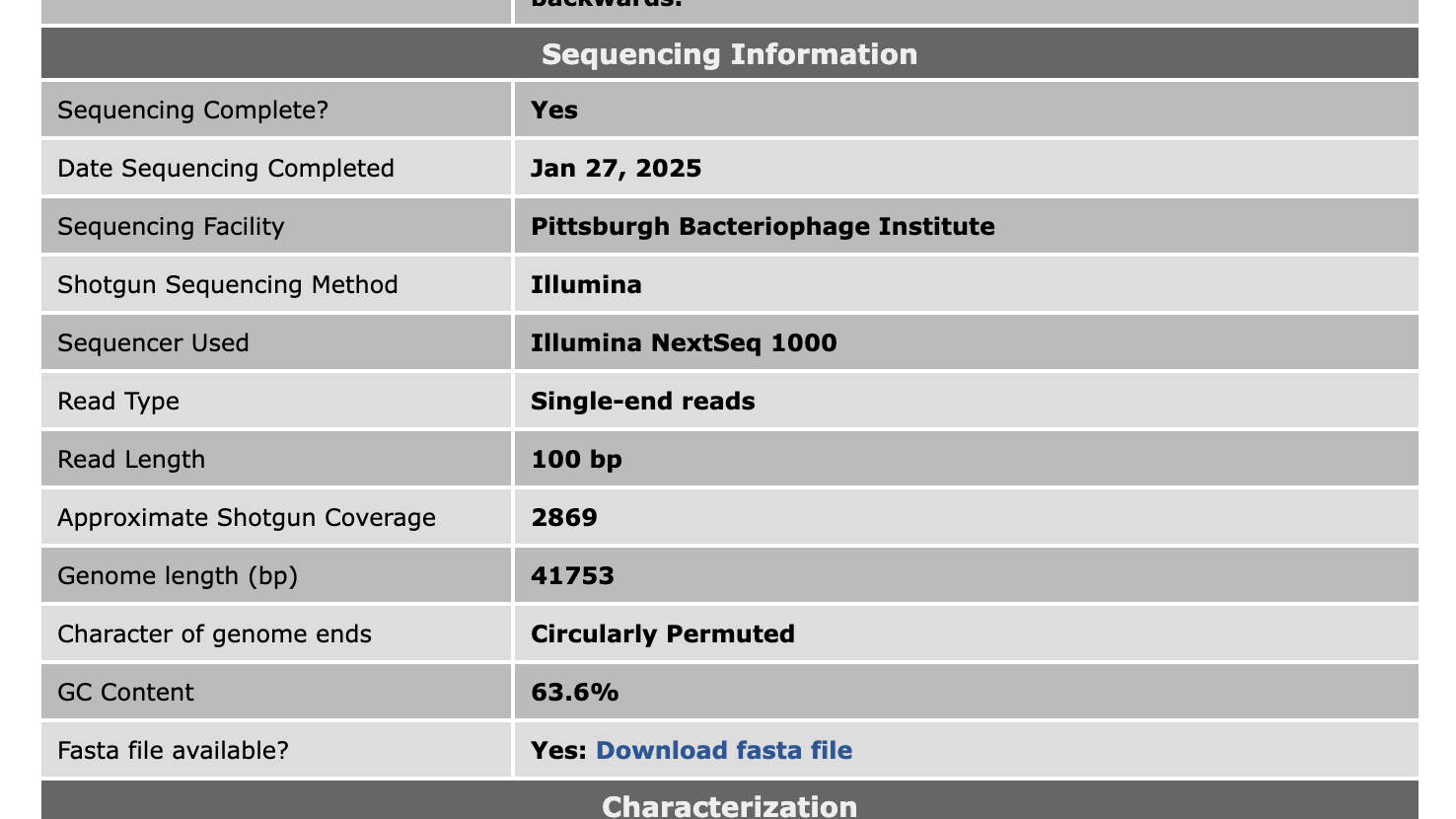 –Dan |
| Link to this post | posted 02 Jan, 2025 16:40 | |
|---|---|
|
|
Hi Fernando, Sounds like your download was probably interrupted at some point, which can happen with these super-big files, and which will cause the machine not to be installable properly. Now, though, with phamerator.org available, we no longer require/support the SEA VM unless there's something specific you'd need it for. Is there something in there you'd still like to use? –Dan |
Posted in: SEA-PHAGES Virtual Machine → SEA VM unzipped size
| Link to this post | posted 31 Oct, 2024 16:58 | |
|---|---|
|
|
Hi Tammy, This is a common question. I'm actually working on updating the PhagesDB database to include type of sequencer, but in the meantime here's a shorthand: If your phage was sequenced by Illumina at Pitt before February 2024, it was on an Illumina MiSeq. If it was March 2024 or after, it was on an Illumina NextSeq 1000. Similarly, if your phage was sequenced on the MiSeq, we recommend adding "Raw" to make your sentence "Raw reads were assembled…" because for phages run on the MiSeq, we haven't done any additional trimming or QC. This will change for the NextSeq 1000 genomes, so if your phage genome was sequenced on that, the reads were trimmed and filtered by: cutadapt 4.7 (using the option: –nextseq-trim 30) and skewer 0.2.2 (using the options: -q 20 -Q 30 -n -l 50) Versions of other programs: Newbler 2.9 Consed 29 For genomic termini, it's too complicated to describe without using most of the 500 words of your MRA, so I usually tell people to just say "as previously described" and cite the chapter I wrote about precisely that. https://pubmed.ncbi.nlm.nih.gov/29134591/ Hope that helps! Oh, and don't forget the "h" in Pittsburgh! –Dan |
| Link to this post | posted 26 Sep, 2024 14:29 | |
|---|---|
|
|
Hi Stephanie, I don't think we ever finalized the sequence of this one, but we do have some Illumina reads which we assembled into WGS contigs. I'll add it to our Nanopore sequencing queue and run it the next time we do a MinION run so we can get the complete sequence. In the meantime, if you'd like to have the contigs we've assembled, I can email those to you. –Dan |
| Link to this post | posted 04 Jun, 2024 17:04 | |
|---|---|
|
|
And another one from Roger Hendrix, from this paper:Comparative examination of [phage] genomes indicates that the hallmark of phage evolution is horizontal exchange of sequences. This is accomplished, first, by rampant non-homologous recombination between different genomes and, second, by reassortment of the variant sequences so created through homologous recombination. |
| Link to this post | posted 04 Jun, 2024 17:02 | |
|---|---|
|
|
I think Graham would probably point out that phages don't really have "lineages" in the traditional sense, which is one of the reasons we don't systematically name them——or use ICTV classifications. Phage genetic material isn't only shared vertically (between different generations), but horizontally as well. Thus, trying to shoehorn phages into classification systems designed for other types of organisms doesn't work well. From a review article Graham wrote in 2008 (this paper): "One of the most striking features of bacteriophage genomes is their apparent mosaic structure; in essence, each genome can be considered as a unique combination of modules that are exchangeable among the population. The size of the modules, their rates of exchange, and the phage genomes carrying them all vary greatly, with phages of different virion morphology, size, and host-range all participants in an orgy of recombination [36]." Hence the advice above. Just explain some version of the above to the reviewer, and let GenBank/ICTV apply whatever designations they see fit! |
| Link to this post | posted 08 Mar, 2024 00:32 | |
|---|---|
|
|
Way back in our "60 phage paper", we grouped phages into clusters based on > 50% nucleotide similarity. Of course, exactly what "50% nucleotide similarity" meant depended on which metrics in particular you looked at, so we used several different ones. You can read about them in the paper. More recently, we've moved to comparing genes rather than nucleotides, and have used ~35% Gene Content Similarity to put phages into clusters. Now, we're probably moving towards using a metric called Protein Equivalence Quotient to cluster phages. It's an ongoing process, and there is no "right" answer and phages will always challenge us by not neatly fitting into bins. PhamClust paper There have been a few groups who have looked at geography, and though there doesn't seem to be any high-level correlation between geography and type of phages found, some subclusters or even genes have only been found in certain geographical areas. –Dan |
Posted in: Phage Biology → Phage Clustering
| Link to this post | posted 13 Feb, 2024 14:57 | |
|---|---|
|
|
eagodin Hi Liz, Just to be clear, the sequences were correct in the sense that all the bases were right. The change was just where Base 1 was, and duplicating a portion of the genome. (Not changing any individual bases or anything.) So everything should be fine/comparable, including the original annotations of those complete ones, but numbers will have changed. –Dan |
Posted in: Phamerator → Cluster FC in Phamerator
