Welcome to the forums at seaphages.org. Please feel free to ask any questions related to the SEA-PHAGES program. Any logged-in user may post new topics and reply to existing topics. If you'd like to see a new forum created, please contact us using our form or email us at info@seaphages.org.
Recent Activity
All posts created by DanRussell
| Link to this post | posted 26 Jan, 2018 20:48 | |
|---|---|
|
|
GregFrederick@letu.edu Hi Greg, To be clear: your problem is with BLAST, not auto annotation itself? Meaning you get all the features, but only some of them have BLAST results? BLASTing all genes can sometimes end up with genes being skipped, especially if done during peak hours. What phage are you working on? –Dan |
Posted in: DNA Master → Auto-annotation fix for fall 2017 and later
| Link to this post | posted 26 Jan, 2018 20:46 | |
|---|---|
|
|
ptsourkas Hi Philippos, Is your DNA Master up to date? If you run Update does all look good? Any other weird errors popping up? I think the servers are working. You've entered the proper access code in the linked document? –Dan |
Posted in: DNA Master → Auto-annotation fix for fall 2017 and later
| Link to this post | posted 22 Jan, 2018 20:04 | |
|---|---|
|
|
Steven Caruso For now! I know the other Steve C. is working on having multiple databases available online, so at some point that will no longer be the case. –Dan |
Posted in: SEA-PHAGES Virtual Machine → Student download of 2016 VM
| Link to this post | posted 22 Jan, 2018 19:59 | |
|---|---|
|
|
Randall DeJong Hi Cal, Did you see my email response? In short: we're not recommending that you install the Virtual Machine at all this year, unless you have a specific need for it. Do you want to proceed with installing anyway? –Dan |
Posted in: SEA-PHAGES Virtual Machine → Student download of 2016 VM
| Link to this post | posted 17 Jan, 2018 17:58 | |
|---|---|
|
|
Hi Claire, I just added a "Last modified" date to the Google Sheet. It should auto-update anytime the list is changed and saved. The date itself is in cell B1. Hope that helps! –Dan |
| Link to this post | posted 17 Jan, 2018 17:56 | |
|---|---|
|
|
I think perhaps our servers were just down around Jan 12. Is it working now? –Dan |
Posted in: DNA Master → Auto-annotation fix for fall 2017 and later
| Link to this post | posted 03 Jan, 2018 16:30 | |
|---|---|
|
|
noverby Hi Nate, There will be no new VM for this academic year. Partly because Phamerator and Starterator reports are now available on the web, we decided that the VM would be optional for schools this year. If they want to, they can use the 2017 VM, but most functions should be available via the web now. –Dan |
Posted in: SEA-PHAGES Virtual Machine → Student download of 2016 VM
| Link to this post | posted 18 Dec, 2017 16:35 | |
|---|---|
|
|
Hi Sarah, From BLASTing Phergie on PhagesDB, it looks like the break is caused by the insertion of a single C. In those other closely-related genomes, there are 4 Cs, but in Phergie there are 5. 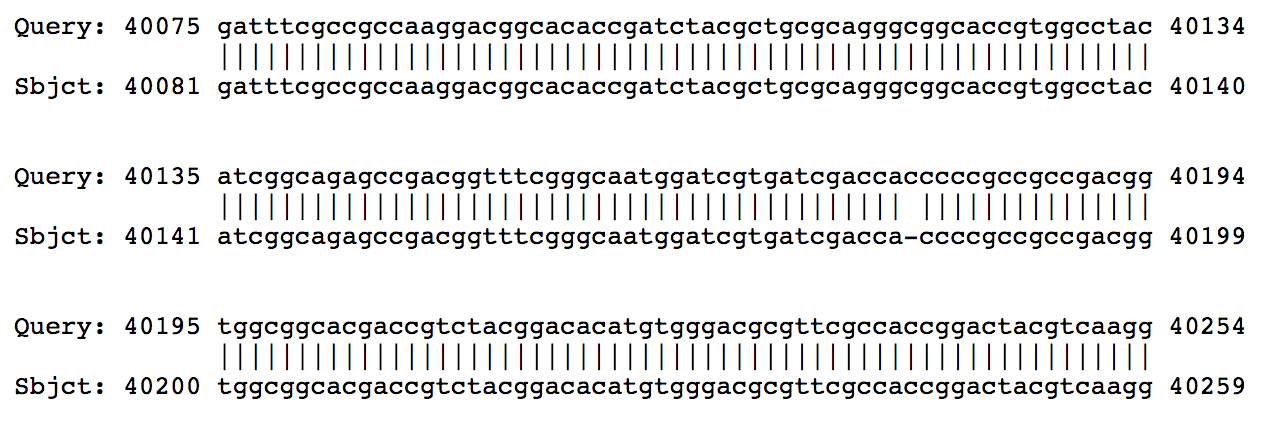 I went and found the assembly for Phergie, and in that region you can see that there are plenty of high-quality reads that all have 5 Cs isntead of 4. 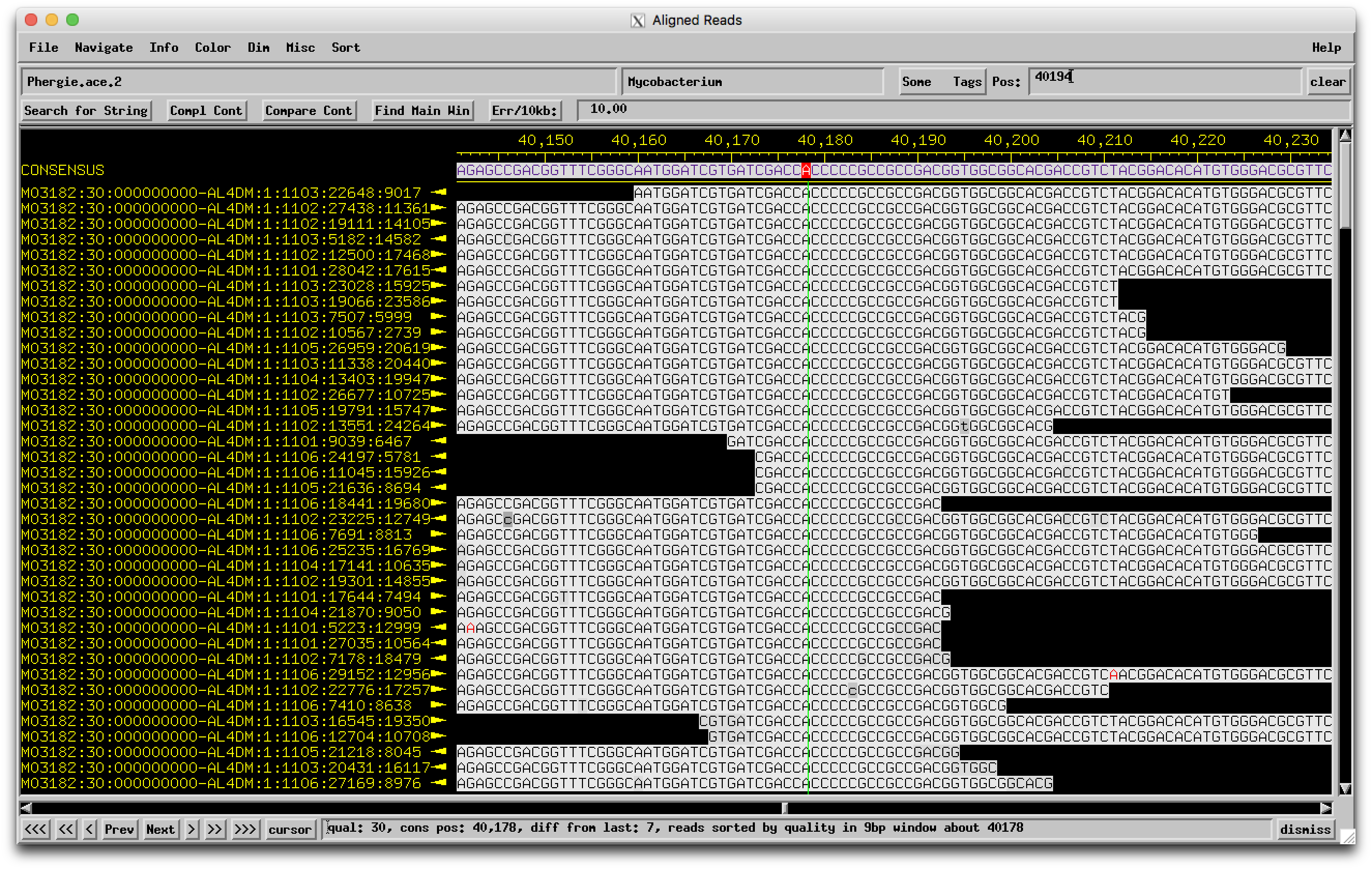 So it's not a sequencing error! Looks like a real mutation that broke that gene into two. –Dan |
| Link to this post | posted 11 Dec, 2017 13:25 | |
|---|---|
|
|
mdgainey Hey Maria, I don't really have any good tips to reduce the host DNA, but my sense is I'd try sequencing it and see what comes out. If the host DNA really is overwhelming, then it's back to the drawing board, but I think there's a decent chance the phage sequence would stand out from the bacterial chunks, and then you'd be done. So I'd just try prepping and sequencing without DNAse and see what comes out, then only worry about reducing the host DNA if that first attempt didn't work. –Dan |
| Link to this post | posted 04 Dec, 2017 16:22 | |
|---|---|
|
|
In 2017, NCBI decided to no longer host public instances of GeneMark and Glimmer, which were used by DNA Master to perform auto-annotations. If your copy of DNA Master is failing to auto-annotate, see the post below about how to get it working again. https://seaphages.org/blog/2017/10/27/auto-annotation-fix-dna-master/ –Dan |
Posted in: DNA Master → Auto-annotation fix for fall 2017 and later
