Welcome to the forums at seaphages.org. Please feel free to ask any questions related to the SEA-PHAGES program. Any logged-in user may post new topics and reply to existing topics. If you'd like to see a new forum created, please contact us using our form or email us at info@seaphages.org.
Recent Activity
All posts created by DanRussell
| Link to this post | posted 30 Oct, 2017 19:57 | |
|---|---|
|
|
Steven Caruso Thanks for the eagle eyes, Steve. I'll let Welkin (the master curator) know. –Dan |
| Link to this post | posted 24 Oct, 2017 15:40 | |
|---|---|
|
|
Hi Katie, I think this error is due to not having a certain library installed on the particular version of Windows that's running. That library wasn't required until after the update earlier this year to use secure NCBI servers. I think I came across that error myself, and in my notes I have that I went to the link below and installed the Visual C++ package there, then restarted and it worked. UPDATE Sep 2021: The link above no longer works, but the file can be found at the link below now. Download it from within Windows and then run it, and make sure you close DNA Master and restart Windows before trying to BLAST again. https://phagesdb.org/static/vcredist_x86.exe Hopefully that helps, –Dan |
Posted in: DNA Master → SSL error
| Link to this post | posted 19 Oct, 2017 16:05 | |
|---|---|
|
|
Hi Heather, As long as everything is precisely typed into Phamerator, I think this is likely to be a Phamerator-Ubuntu issue rather than a PhamDB issue. Check out the suggestions in this post and see if any of those might help. Sometimes, weird stuff can happen on the VM which sends Phamerator into strange cycles, but Steve had a couple suggestions. https://seaphages.org/forums/topic/246/?page=1#post-1338 –Dan |
Posted in: Phamerator → PhamDB: Make your own Phamerator databases
| Link to this post | posted 29 Sep, 2017 14:16 | |
|---|---|
|
|
grosej Hi Juli, I don't know if you can merge two databases easily with PhamDB, but PhamDB definitely makes it easy to add phages that are in the system to other databases in the system without having to re-enter them. You can use the "Add to Database" function. –Dan |
Posted in: Phamerator → Phams
| Link to this post | posted 03 Jul, 2017 15:42 | |
|---|---|
|
|
Hmmm…I was totally ready to say Chris is right, it's probably a 454 sequencing error, but here is the region in question in the assembly: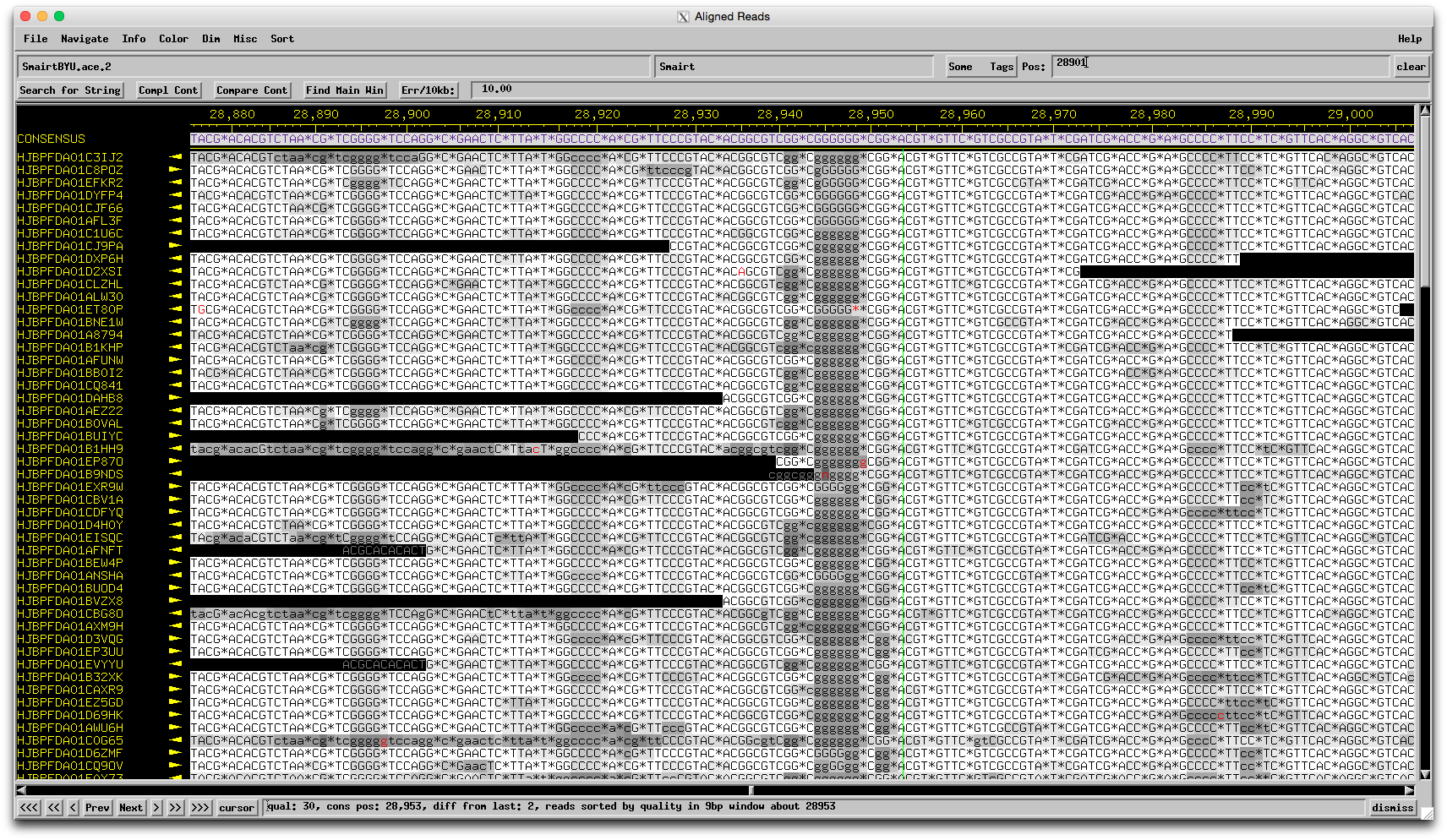 The evidence for 6 Gs there is quite strong, and there are only a few reads that suggest only 5 Gs. That doesn't mean it's not truly 5 Gs, but I wouldn't feel comfortable changing the sequence on this evidence alone. I guess that means you should proceed as is? What do you think Welkin/Chris? –Dan |
Posted in: Gene or not a Gene → Gene split in 2
| Link to this post | posted 25 Apr, 2017 19:26 | |
|---|---|
|
|
Summer Research Opportunity Next-generation Fluorescent Reporter Mycobacteriophages The Jacobs laboratory has innovative and multifaceted studies on the biology of Mycobacterium tuberculosis and its phages. We are seeking undergraduates who have taken the SEA-PHAGES course to work on an exciting project to generate next-generation fluorescent reporter mycobacteriophages. We have previously shown that fluorescent reporter mycobacteriophages can be used to rapidly assess drug susceptibilities of Mycobacterium tuberculosis cells in culture or directly from sputum samples (Jain et al., 2012 Journal of Clinical Micro. 50:1362-9). Recently, we have developed dual reporter mycobacteriophages that can be used to identify M. tuberculosis persister cells, a subpopulation of cells in a culture that are drug tolerant (Jain et al., 2016 mBio. 7 e01023-16). The summer program will focus on generating improved versions of reporter mycobacteriophages (greater sensitivity, shelf-life etc.). This experience will help students explore nuances of molecular biology and the scientific method. Students with a basic background in biology, recombinant DNA technology, and phage biology would be ideally suited for this project. The students will not need to work with virulent M. tuberculosis strains as we have developed Biosafety Level 2 strains for this project. The Jacobs Lab is located in the Price Translational Research Center at the Albert Einstein College of Medicine, 1301 Morris Park Avenue, Bronx, NY. Einstein is near the Bronx Zoo and the New York Botanical Gardens (both ideal phage hunting areas!). The position will come with an stipend of $11,000 for 3 months. If interested, please contact Dr. William R. Jacobs at jacobsw@hhmi.org Lab webpage: http://williamrjacobs.org |
| Link to this post | posted 24 Apr, 2017 16:18 | |
|---|---|
|
|
Hi Sangha, From looking at the DNA Master file, I think that the longer of these two (which overlaps a lot of other tRNAs) is just a miscall. You can safely delete that one. The 425 bp one, however, is probably a tmRNA, and should be called. –Dan |
Posted in: Gene or not a Gene → C1 phage
| Link to this post | posted 12 Apr, 2017 14:38 | |
|---|---|
|
|
Shallee Page Hi Shallee, This happens from time to time. Looks like the server has been reset and is working now. –Dan |
Posted in: DNA Master → DNA Master failed update
| Link to this post | posted 04 Apr, 2017 17:44 | |
|---|---|
|
|
Hi Nancy, These are probably tRNA genes, which don't appear in Phamerator (yet!). So for those you'll have to follow some of the tRNA guidelines rather than Starterator, Phamerator, etc. –Dan |
Posted in: Phamerator → Missing Genes in Phamerator Ading_draft
| Link to this post | posted 28 Mar, 2017 14:29 | |
|---|---|
|
|
Hi Victoria, If Phamerator ever prompts you for a password, it should just be: "phage". If that doesn't work, something else has gone wrong! –Dan |
