Welcome to the forums at seaphages.org. Please feel free to ask any questions related to the SEA-PHAGES program. Any logged-in user may post new topics and reply to existing topics. If you'd like to see a new forum created, please contact us using our form or email us at info@seaphages.org.
Recent Activity
Lysin A split in 2 or sequencing error-B1 cluster
| Link to this post | posted 14 Dec, 2017 17:03 | |
|---|---|
|
|
I have come across an anomaly while trying to wrap up the annotation QC of a B1 phage, Phergie. gp47 and gp48 both have BLAST hits to LysA. When comparing this area to other very similar phage, there is one longer ORF in this position that is called as LysA. There is strong coding potential for both ORFs but the reading frame shifts. I do not know enough about sequencing and finishing genomes to tell if this is just an error (which is my initial thought) or something more interesting. Thoughts? |
| Link to this post | posted 18 Dec, 2017 16:35 | |
|---|---|
|
|
Hi Sarah, From BLASTing Phergie on PhagesDB, it looks like the break is caused by the insertion of a single C. In those other closely-related genomes, there are 4 Cs, but in Phergie there are 5. 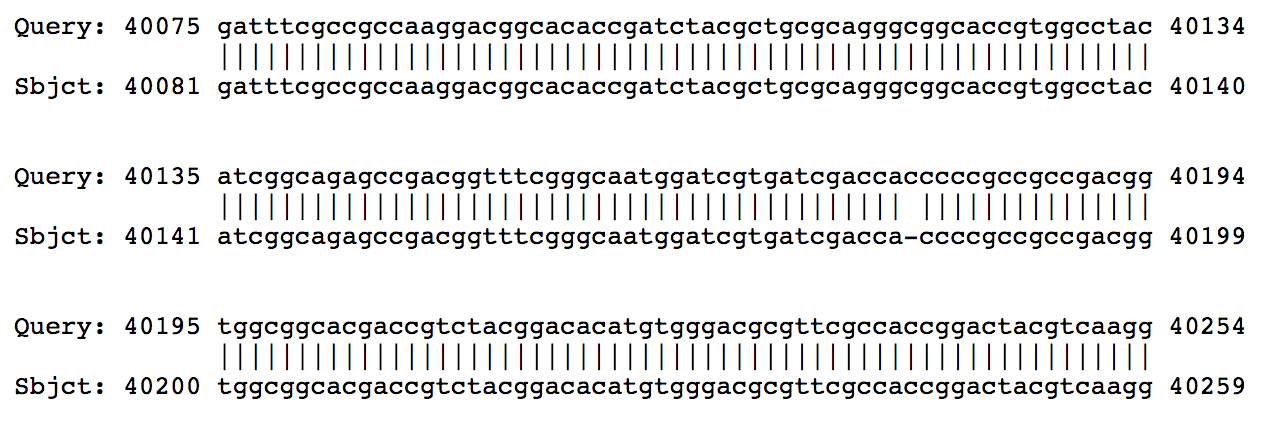 I went and found the assembly for Phergie, and in that region you can see that there are plenty of high-quality reads that all have 5 Cs isntead of 4. 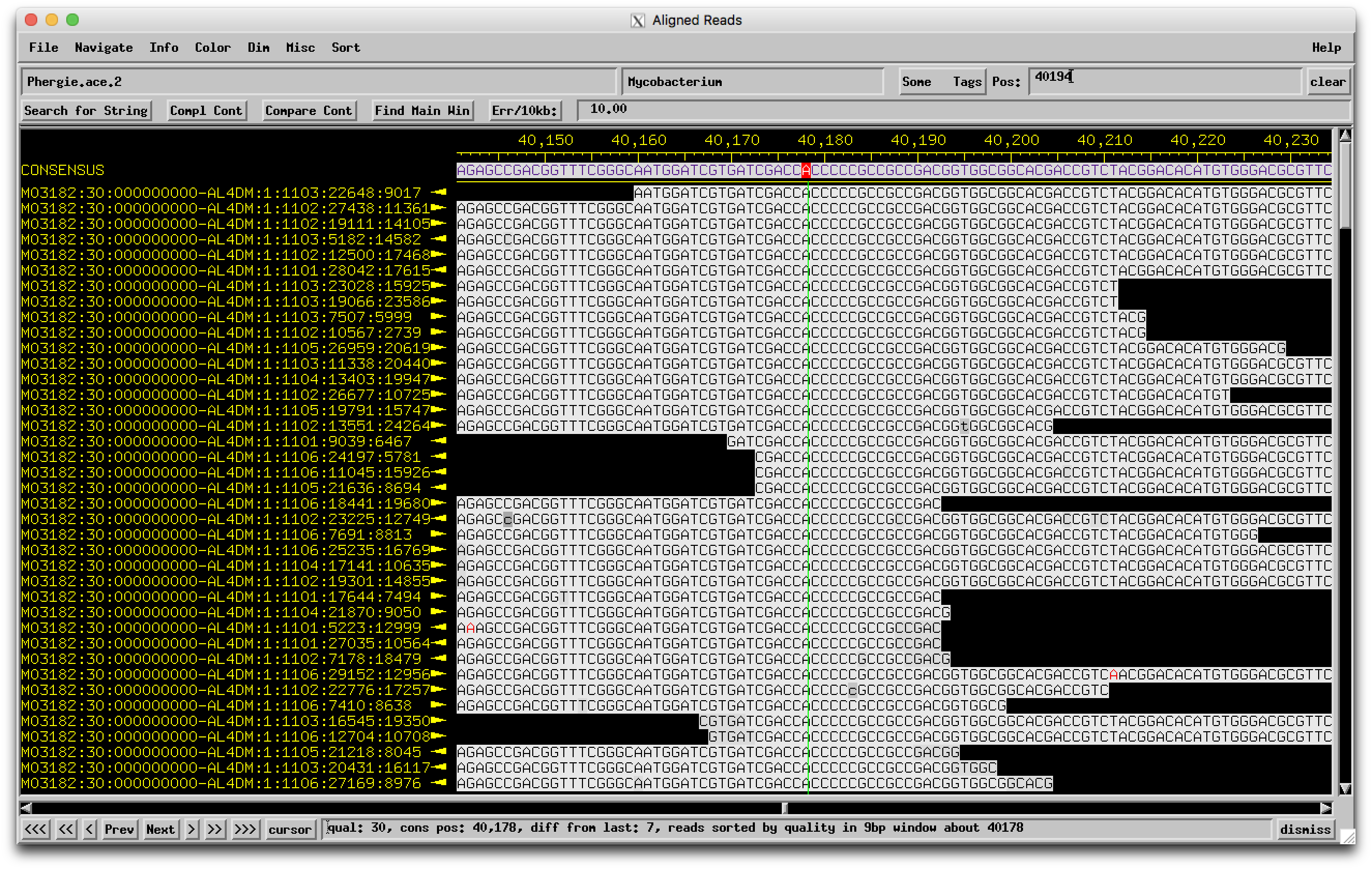 So it's not a sequencing error! Looks like a real mutation that broke that gene into two. –Dan |
| Link to this post | posted 18 Dec, 2017 16:51 | |
|---|---|
|
|
Cool! Sarah, check to see if it is split into two functional domains like in some of the Gordonia phages. This would be the first example of this in the Mycobacteriophages. |
| Link to this post | posted 18 Dec, 2017 18:12 | |
|---|---|
|
|
It looks like each gene gets a strong conserved domain hit in BLASTp (see attached screen shot). So should I list the functions as "lysin A, peptidase domain" and "lysin A, amidase domain"? |
| Link to this post | posted 19 Dec, 2017 17:47 | |
|---|---|
|
|
I wonder if the poly-C is conserved in the Gordonia lysins… |

 75Kb
75Kb