Welcome to the forums at seaphages.org. Please feel free to ask any questions related to the SEA-PHAGES program. Any logged-in user may post new topics and reply to existing topics. If you'd like to see a new forum created, please contact us using our form or email us at info@seaphages.org.
Recent Activity
Dan Russell posted in Congrats to Steve Caruso and Beth Wilkes — 2025 ASM Outstanding Instructor Award, Honorable Mention
ACMPhageHunters posted in Clarification Question About HNH Endonuclease Function Determination in view of hits to the Ref Sequences
cdshaffer posted in Clarification Question About HNH Endonuclease Function Determination in view of hits to the Ref Sequences
All posts created by scaruso
| Link to this post | posted 11 Mar, 2019 19:03 | |
|---|---|
|
|
Claire, Thank you very much for the response, that was very helpful. And I can delete Anthony now, we are done with it. Though the answer will, no doubt, be useful for other duplicates in the future. Steve |
| Link to this post | posted 09 Mar, 2019 22:07 | |
|---|---|
|
|
Claire, Can you tell me what the Phagesdb Function Frequency is based on? I see a list of various function assignments for a variety of PHAMS, but what brings them up? I don't see the same hits on Blastp or HHPred, always, and the PHAMS are different than the gp being examined, so I'm not clear. By the way, I posted another question about HHPred results in PECAAN and external, any thoughts about that? Thanks! Steve |
| Link to this post | posted 05 Mar, 2019 19:12 | |
|---|---|
|
|
Excellent. That's very helpful. Thank you! Steve |
| Link to this post | posted 05 Mar, 2019 03:21 | |
|---|---|
|
|
A follow up on this old post, if you don't mind. There is a recent post in the Cluster Specific Tips that continues this conversation. See: Forum Home / Science / Cluster-Specific Annotation Tips / Cluster A Annotation Tips / minor tail proteins Welkin Pope posted 03 Apr, 2018 14:11 "Unlike many other clusters, the Cluster A phages have *some* minor tail proteins at the left end of their genome, upstream of the lysins and terminase genes (around gene 4-6ish). You can recognize these proteins due to their size. Some of them may have structural motifs that suggest long, extended proteins, like collagen-repeats, or coiled-coils." So I would like to get a little feedback if I can. Heather gp18 (a BB2) has BlastP hits to lots of "minor tail protein" calls, as well as some "tail fiber" and other similar calls. But I don't think they are justified. HHPred actually hits to collagen, not to any identified tail proteins. I think it's a rabbit hole. And then there's: The Collagen-like Protein gp12 Is a Temperature-dependent Reversible Binder of SPP1 Viral Capsids. Mohamed Zairi, Asita C. Stiege, Naima Nhiri, Eric Jacquet, and Paulo Tavares http://www.jbc.org/content/289/39/27169.full Which shows that these hits might be for other structural components, like parts of the capsid. My inclination is to leave it NKF, unless I am missing something. Any thoughts? >Heather_gp18 MTQPVETFPLPPSIETVKVHGHYRGPDGRGLQGTVTFTGPGLLTFPDADLFIAGPVVARLDEFGAFEVTLPATDNEGMNPSDWSYTVKENL TGVTGARTFALLLPKDTAEIDLADVAPADPLTPNYVPVPGPQGEPGPQGVKGDPGATGPAGANGAPGAPGAAGAPGVIQSVNGKSAASVTL VPADLGAVPTSDKGVANGVASLGADGKVPAGQLPATSNAVTSVNTKTGAVVLSASDVSAVPTSDKGAANGVATLDASTKVPTAQIPSLTST YVAVSTRGAANGVATLDGTTRLPVAQVPATLPKNTWTPQALGFQAWSCDPGGVANPGAKYLTPQRLYMTGFNITEPTTVSNVVMFARGYGG VGTNRYMAGIYKEDGTRVVASAAVALSMAGQESGALPGMVTNHIGAVPLPITSTTLQPGRYWVAWVLTVGAASDFAFYHVQNEAPVSTANF FMPTTPFARAWYLAGQSTLPTTVSQTAAGVLADHDIPIVALA |
| Link to this post | posted 01 Mar, 2019 19:53 | |
|---|---|
|
|
I think you have a clear holin already as well, Esketit_Draft gene 30 (25259 - 25618 ), and it's the same length as the other BI1s (119 aa) and similar to the BI2 Scap1 that has it as well (129 aa). I don't know if we have confirmed cases of two holins (Lee?). I would be hesitant to call a second one, I think. Steve |
| Link to this post | posted 15 Feb, 2019 13:41 | |
|---|---|
|
|
Sounds good. Thanks! Steve |
| Link to this post | posted 15 Feb, 2019 12:53 | |
|---|---|
|
|
We would like to add the gene function “DNA delivery” (or equivalent wording to that effect) to the official functions list. This is the function assigned to the Tectivirus Enterobacteria phage PRD1 p32 and p34. It is also a function commonly assigned in Tectiviridae. Please see: Jalasvuori M, Koskinen K. 2018. Extending the hosts of Tectiviridae into four additional genera of Gram-positive bacteria and more diverse Bacillus species. Virology 518:136–142. PMID: 29481984 We identified this function in Streptomyces phage Forthebois gp22 and gp28 by HHPred to PRD1 p32 and p34. The functions of PRD1 p32 and p34 are described here: Grahn AM, Daugelavicius R, Bamford DH. 2002. The small viral membrane-associated protein P32 is involved in bacteriophage PRD1 DNA entry. J Virol 76:4866–72. PMID: 11967303 “The amino acid sequence of P32 shows characteristic elements for integral membrane proteins. According to the orientation of the predicted ɑ-helix, the topology of protein P32 would be such that the N-terminal part spans the membrane, leaving the bulk of the protein (C terminus) in the cytoplasmic side of the CM. If our model of a clathrin-like engulfment of the phage membrane from the host cytoplasmic membrane during particle assembly is valid, the C terminus of P32 would face outward on the virus membrane surface… Sequence analysis revealed another interesting feature, a close similarity of P32 to another small phage membrane protein, P34 (Fig. 5). This is a second example of a high level of sequence similarity between two PRD1 gene products, suggesting a single gene origin for the two elements.” Saren A-M, Ravantti JJ, Benson SD, Burnett RM, Paulin L, Bamford DH, Bamford JKH. 2005. A snapshot of viral evolution from genome analysis of the tectiviridae family. J Mol Biol 350:427–40. PMID: 15946683 “The known PRD1 genes encode replication (P1, P vertex and injected into the host cell through a tubular structure derived from the viral membrane.” “High conservation occurs in the DNA polymerase encoding sequence (gene I), except close to the gene end. This is also the case with the region encoding the carboxyl terminal end of the packaging ATPase (gene IX), the regions coding for the small membrane protein linking the packaging vertex to the membrane (gene XXII), the coat protein (gene III), and the small membrane proteins important for DNA delivery (genes XVIII, XXXII, XXXIV).” >PRD1_p32 MGEFGKTLITIVTAIIGVAIIAVIVSQRSNTAGVIQSATSGFSNILKSALAPII >PRD1_p34 MNDFVGPIVTVLTAIIGVAILAVLVSRNSNTAGVIKAGSGGFSSMLGTALSPVTGGTGFAMTNNYSGF >Forthebois_gp22 MNGDKVFNVLGAIVTVALVTTIVSRPTSAQVIKAMGDAFSGSIRAALGKZ >Forthebois_gp28 MKGDDVIKVLMGIIAVALVTTIVMRPNSATVIKAAGSAFSGSLRAAMGKZ Forthebois gp22 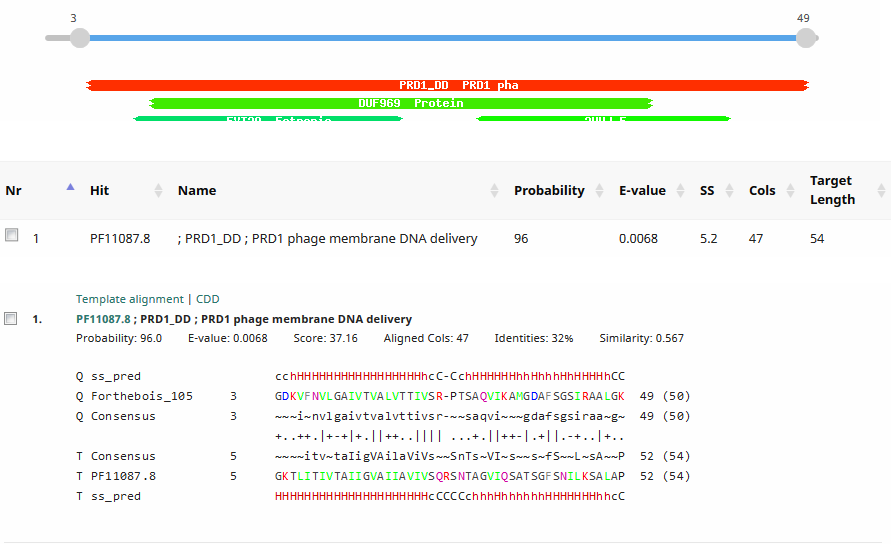 Forthebois gp28 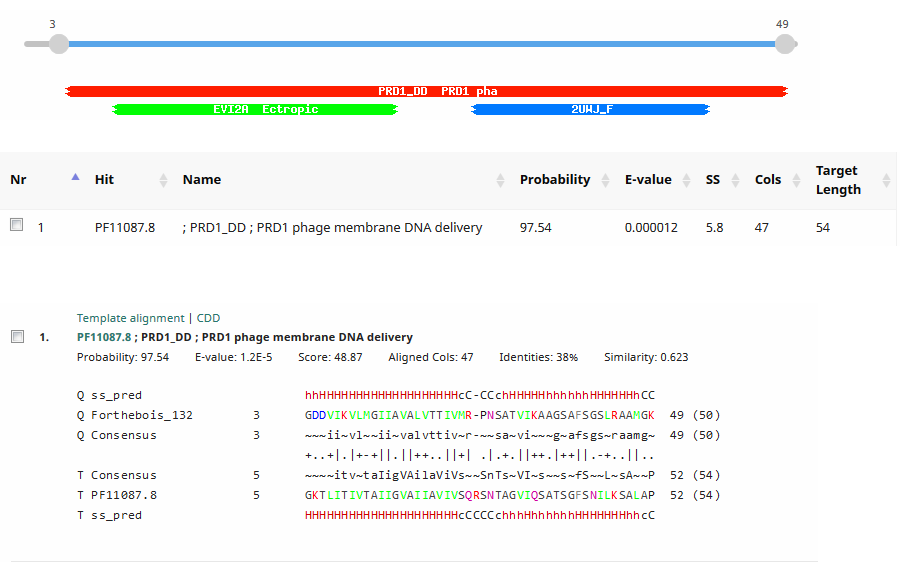 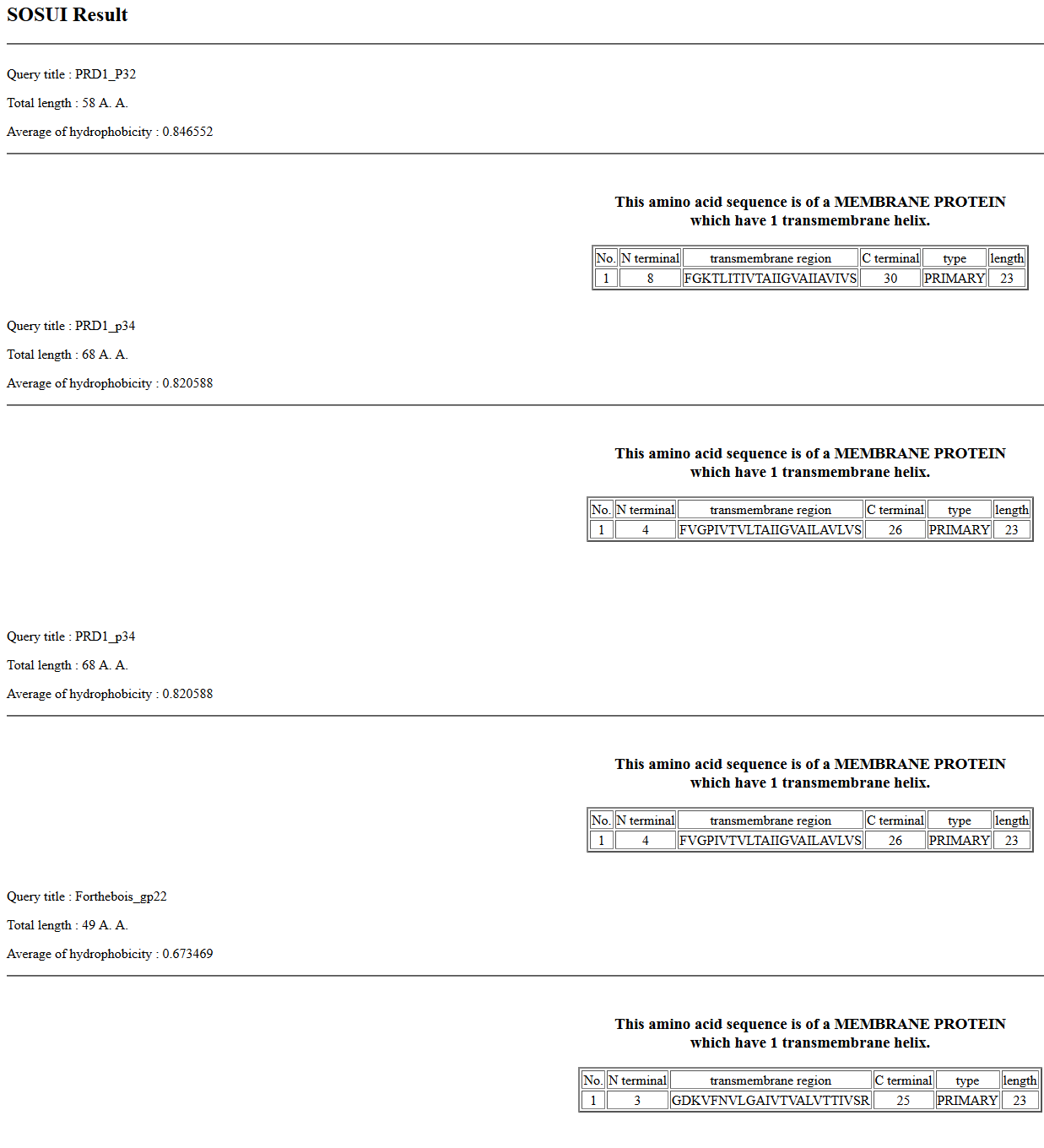
|
| Link to this post | posted 27 Nov, 2018 17:47 | |
|---|---|
|
|
Thanks, Debbie. I appreciate you looking at it. Steve |
| Link to this post | posted 27 Nov, 2018 01:33 | |
|---|---|
|
|
jparkerSteven Caruso Not that we are aware of yet. We are still using the old fashioned one for Bacillus phages for now, though we eagerly anticipate a web-based version for the students. There is a benefit to having access to the old version, though, if you want to be able to pull pham data for analysis, though, for either family. Steve |
| Link to this post | posted 24 Nov, 2018 15:22 | |
|---|---|
|
|
I was thinking Synteny might be compelling, but I ran the gene with the other databases in HHPred, here's what I find: TIGR01673 holin_LLH; phage holin, LL-H family. This model represents a putative phage holin from a number of phage and prophage regions of Gram-positive bacteria. Probability: 91.2 E-value: 2.9 Score: 27.85 Aligned Cols: 83 Identities: 14% Similarity: 0.192 The second gene's (24921) best hit is to an uncharacterized membrane protein in the host seen by mass spec. NP_217999.1 membrane protein [Mycobacterium tuberculosis H37Rv] Probability: 99.07 E-value: 1.1E-12 Score: 92.87 Aligned Cols: 77 Identities: 30% Similarity: 0.671 So, seems like a reasonable call, but I can understand if you want to let it go. As for the other gene I mentioned above, I can't find any reason to call () an excise or DNA binding protein other than that it was called that by someone. So I'm going to call it NKF, though if you have info I would be grateful for it. Otherwise, it's all done. You can see the whole thing in PECAAN (or on a DNAM) if you like for reference. Thanks, Steve |

