IMPORTANT DATES
March 24 - 28, 2025: Registration Window
May 16, 2025: Deadline for abstract submission
May 23, 2025: Those selected to give talks will be notified.
May 29, 2025: Deadline to upload posters to be printed by HHMI
June 6 - 9: Attend SEA Faculty Meeting
WHO SHOULD REGISTER?
HHMI HQ has a meeting capacity of 175 persons. Given the size of our community, this means that we invite 1 faculty member to represent each SEA institution. An overview of the agenda is provided below to help you decide who best to represent your institution. You may nominate a second faculty member to attend, in-person, in particular for schools offering both the PHAGES and GENES projects. An invitation will be extended to the second person based on space availability.
HOW TO REGISTER
A registration link was sent by email to all SEA faculty on March 24th. HHMI Travel Services will use this registration information to contact participants and begin arranging travel.
WHAT IS ON THE AGENDA?
Final details of the agenda are still being developed. Here is some helpful information for now.
- Guests for the Faculty Meeting can expect to arrive at HHMI Headquarters by 3:00 PM EDT on Friday, June 6, and the Meeting will conclude by 12:00 PM EDT on Monday, June 9.
- There will be a session for faculty posters and talks. As you submit your abstracts, you will have an opportunity to indicate if you would like your abstract considered for a talk.
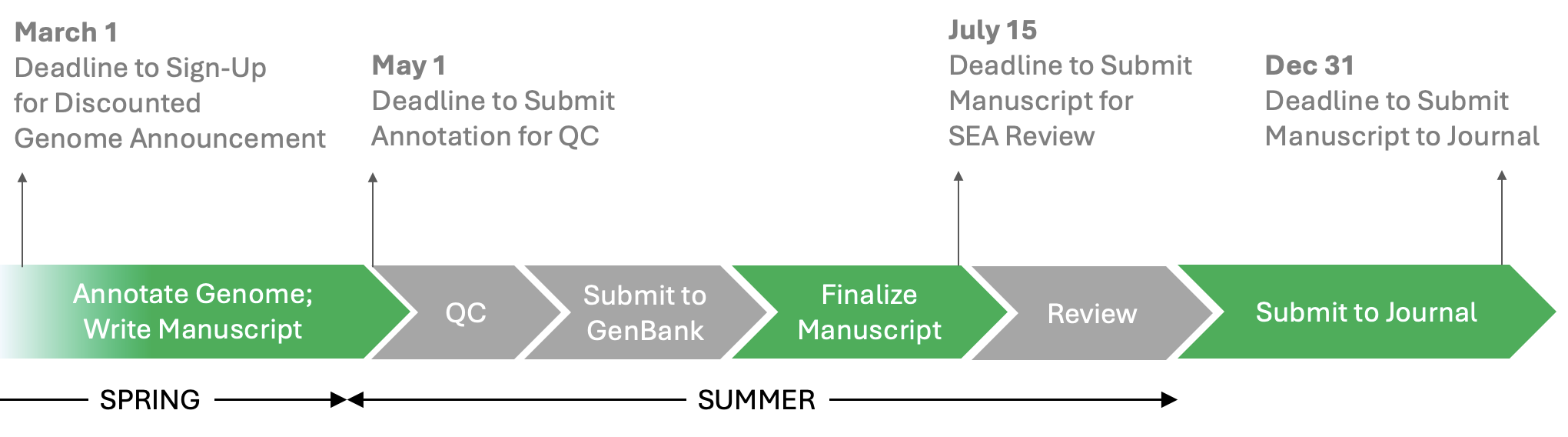
- The meeting will primarily feature working sessions to improve our collective work, including to improve our annotations (e.g. cluster-specific annotation work), to advance our understanding of research tools and methodologies (e.g. phage clustering, Alpha-fold, lysogeny), to write reports and manuscripts for publications (e.g. genome announcements, GENES reports, Cluster Reports), and to engage in science pedagogy research projects. When registration opens, you will have an opportunity to suggest additional topics or sessions you would like to have included at the meeting.
HOW TO SUBMIT AN ABSTRACT ?
As a way for all of us to share and discuss ideas and data, we encourage you to present a poster or a talk. Posters can FOCUS on one brief item (e.g. idea, question, data point for discussion) or a more developed or even published story. They can be on a range of topics, from intriguing experimental data to pedagogical resources. Submit your abstract using the link at the top of your institutional page on seaphages.org. If you would like your abstract to also be considered for a talk, you can indicate that during abstract submission.
WHAT ARE POSTER SPECIFICATIONS?
Posters should be 3-feet wide. They can be up to 4-feet tall.
WILL HHMI PRINT THE POSTER?
If you would like your poster printed for you, upload your poster >> HERE << by May 29. HHMI will have your poster printed and shipped directly to HQ.
WHAT WILL IT COST?
HHMI will cover the cost of travel to and from the meeting, and provide lodging and meals throughout the meeting.