Welcome to the forums at seaphages.org. Please feel free to ask any questions related to the SEA-PHAGES program. Any logged-in user may post new topics and reply to existing topics. If you'd like to see a new forum created, please contact us using our form or email us at info@seaphages.org.
Recent Activity
Putative Frameshift in a Reverse gene in N-Cluster Phage BabeRuth
| Link to this post | posted 17 Nov, 2018 21:42 | |
|---|---|
|
|
As part of the University of Western Carolina Hackathon (Funathon!), we annotated the genome of the N-Cluster phage BabeRuth. ORFs 33 and 34 in the draft BabeRuth genome both hit to ORF32 in the annotated Redi genome in the exact way you would expect for a frame-shift. If you zoom into the nucleotide region around 26445 in BabeRuth and align it with the homologous region in Redi, you will see that there is a one bp insertion in BabeRuth that came out of the sequencing. Redi has the coding sequence 5'-CTGGGGCG and BabeRuth is 5'-CTGGGGGCG. So, the difference in sequence is in a poly-G tract, a sequencing no man's land in some technologies. Interestingly, there is an amino acid sequence in this region that is N-DGAAGAAD in Redi's protein product. Don't know if this has any significance. Redi was sequenced in 2011 (454? Technology not noted in PhagesDB) and BabeRuth in 2017 on Illumina. All of the other N-cluster genomes with this ORF were sequenced on Illumina as well and they have a single ORF like Redi. At this point, we are not sure if this has arisen from a sequencing issue in BabeRuth. Is it worth doing Sanger sequencing on BabeRuth to look at that region of the genome? Alternatively, should we just call ORF 33 in BabeRuth as a truncated version of ORF32 in Redi? Thanks for any insights people can provide! |
| Link to this post | posted 19 Nov, 2018 17:51 | |
|---|---|
|
|
Hi Bill, Since BabeRuth sounds like the outlier here, I went back and took a look at the BabeRuth sequencing data. Here's a pic of that region. 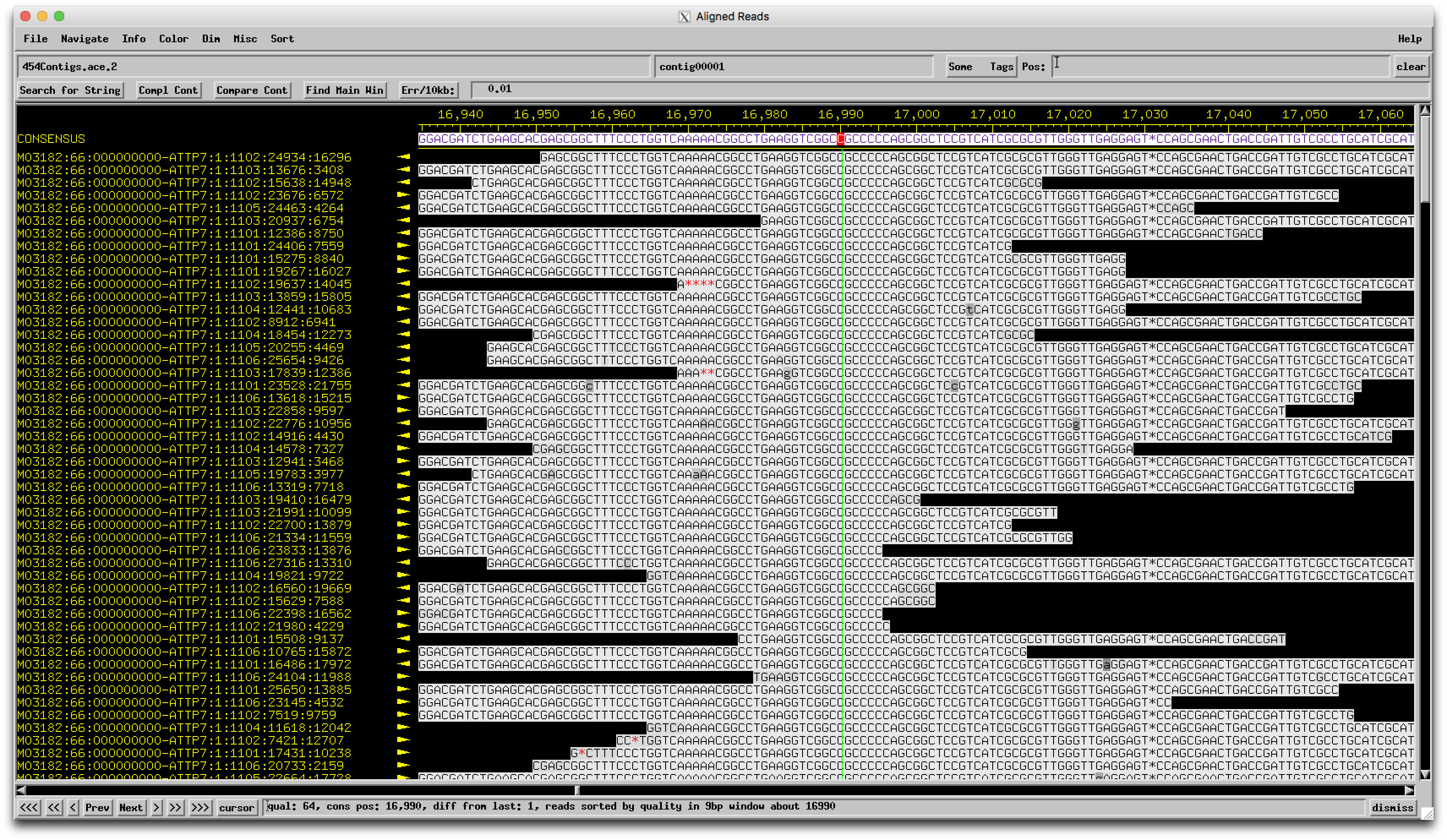 Very solid coverage and agreement that there are indeed 5 Cs here, not 4 like the others with similar genomes. This is a real biological feature, not a sequencing error! I can't say for sure of course whether this is really a frameshift (in which case the protein would still be made) or just a mutation that truncates the protein (more likely), but the underlying DNA sequence at least is correct. Good eye, and this is definitely the kind of thing that's worth checking. –Dan |

