There are two primary pieces of software needed during the second semester of the SEA-PHAGES program.

DNA Master

Phamerator
There are also a number of other programs that can be used in the SEA-PHAGES program.

SEA Virtual Machine

SEA-PHAGES App

Newbler
Consed

AceUtil

Starterator
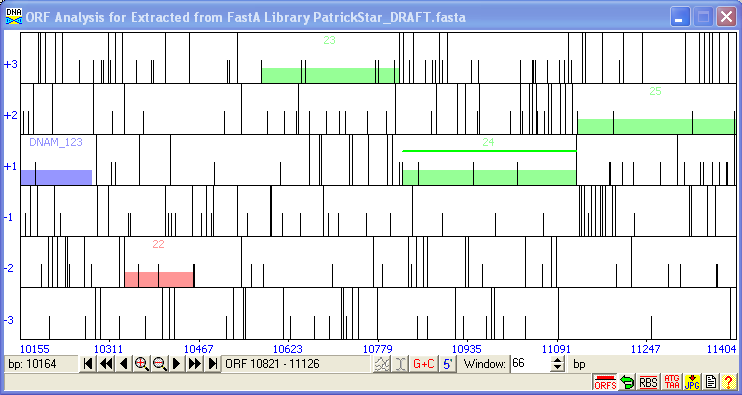
In the SEA-PHAGES course, DNA Master is used for annotating and analyzing the phage genomes discovered by students. DNA Master allows students to quickly generate an auto-annotation using GeneMark and Glimmer to make gene calls and Aragorn and tRNAscan-SE to find tRNAs. They can then refine the auto-annotation calls by investigating evidence for alternative start sites, identifying missed or erroneously called genes, and determining gene functions.
DNA Master runs ONLY on Windows machines, though it can run within a Windows virtual machine on a Mac. You can find information and an installation guide here.
A guide to using DNA Master for phage genome annotation is available here.
Info on troubleshooting common issues can be found here.
Questions? Visit our DNA Master forum.
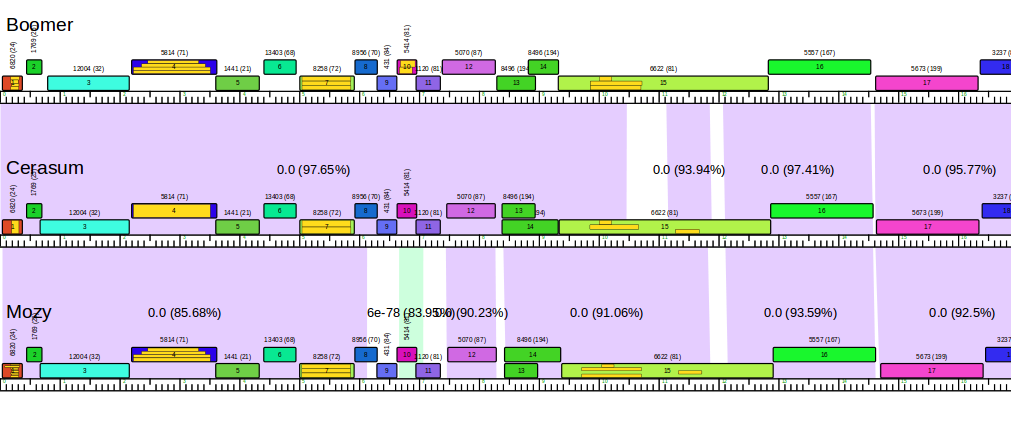
Phamerator is a comparative genomics and visualization program that allows SEA-PHAGES students to compare their genomes with the larger collection of Actinobacteria phage genomes. It compares the amino acid sequences of all genes in a collection of phages and groups similar genes into Phamilies (aka Phams). Users can then view, compare, save, and/or print color-coded genome maps.
Historically, Phamerator ran only on Linux. Fortunately, it is now available on the web at phamerator.org. We strongly recommend you use Phamerator on the web for analyzing your SEA-PHAGES genomes.
If you need access to the Linux version of Phamerator, it is available as part of the SEA Virtual Machine.
Questions? Visit our Phamerator forum.
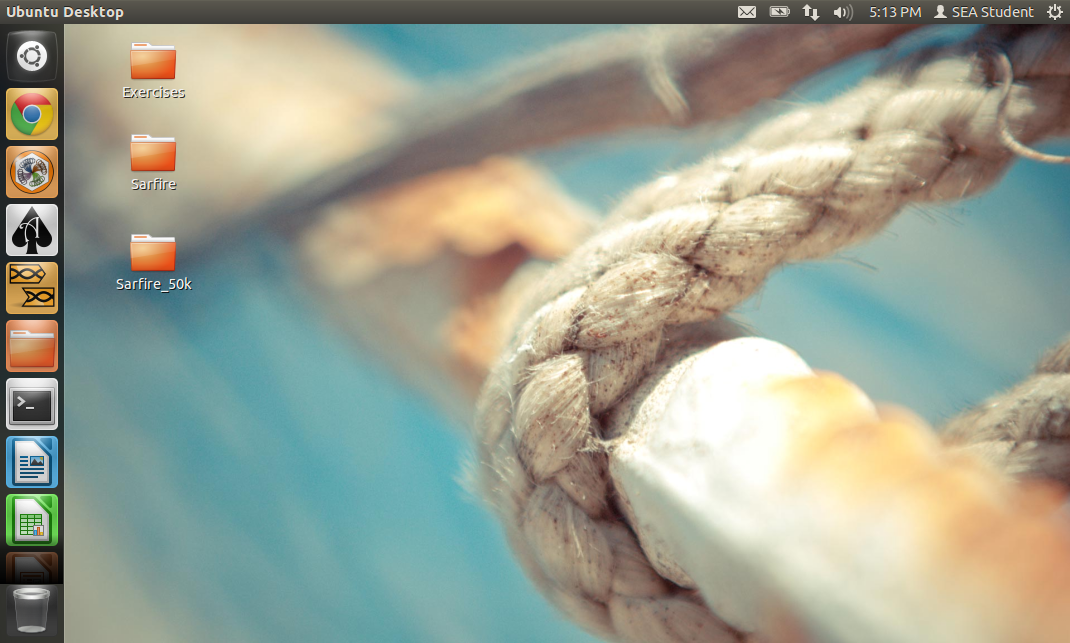
The SEA-PHAGES Virtual Machine is a pre-made Linux (Ubuntu) image that has a number of useful programs pre-installed, including (legacy) Phamerator, Newbler, Consed, AceUtil, and Starterator. Using a virtualization program like VirtualBox, it can be installed on Windows or Mac computers.
If you are a SEA-PHAGES faculty member, you can download the SEA Virtual Machine here.
Questions? Visit our SEA-PHAGES Virtual Machine forum.
The official SEA-PHAGES App is now available for iOS and Android devices. It can also be accessed by computer. The app serves three major functions: a social network for all SEA-PHAGES participants, a way to keep track of your soil sampling locations, and a player for the Sounds of the SEA podcast.
The App is available from the Apple App Store or Google Play.
Questions? Visit our SEA-PHAGES App forum.
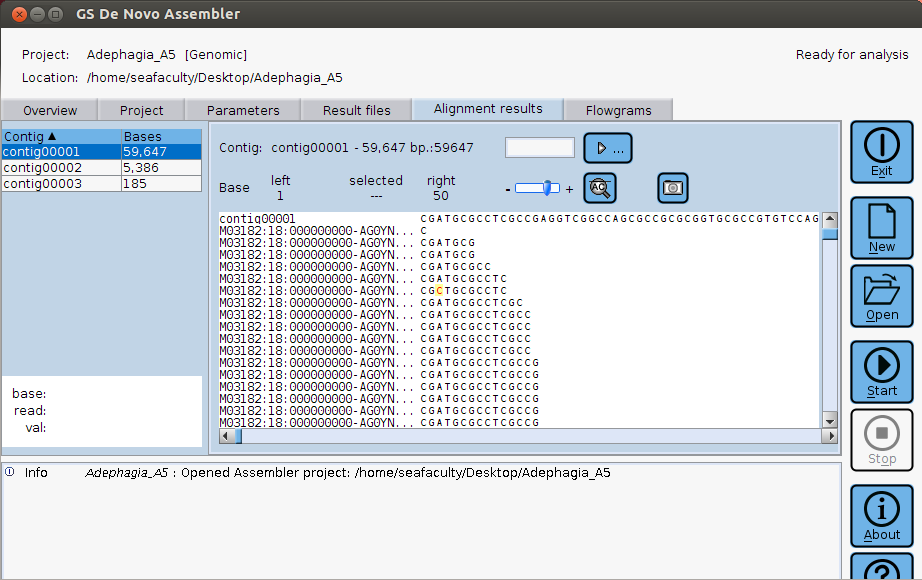
Newbler (more formally: GS De Novo Assembler) is a de novo assembly program that can be effectively and quickly used to assemble phage genomes sequenced by Illumina, 454, or Ion Torrent technologies. Sequencing reads can be in .fastq or .sff format.
Some tutorials for how to use Newbler are available on PhagesDB.
Newbler runs ONLY on Linux. It is available as part of the SEA Virtual Machine.
Questions? Visit our Newbler forum.
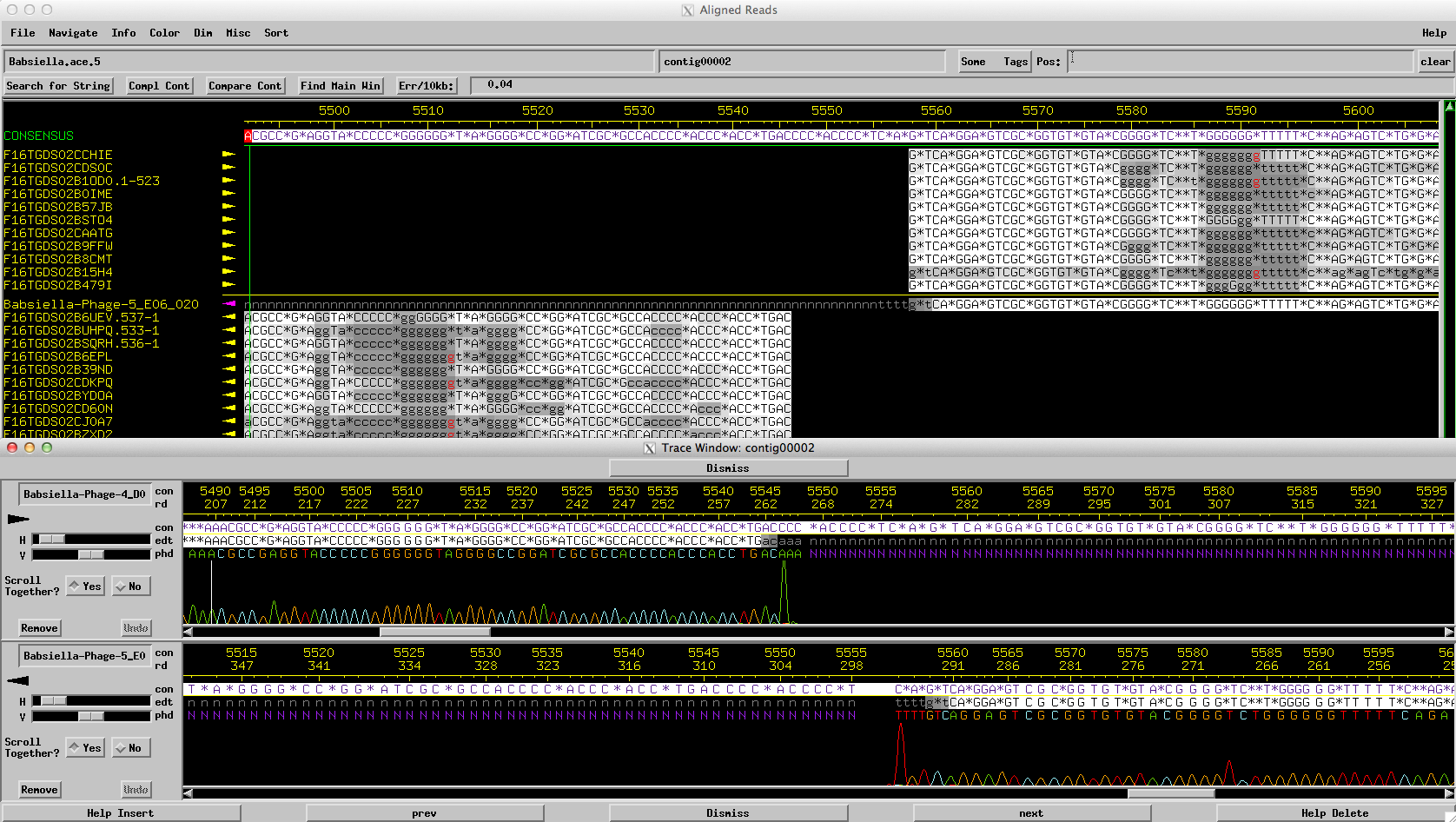
Consed is a program that allows the user to view, edit, and finish assembled genome sequences. It is used in the SEA-PHAGES program by members of the GIFT to ensure that the phage genome sequences returned to schools are complete and accurate. Many schools have begun using consed to finish their own phage genomes, and it can be used as an educational tool to show students how their final genome sequence was determined from shotgun sequencing reads.
Some tutorials for how to use consed are available on PhagesDB.
Consed is available as part of the SEA Virtual Machine, or can be acquired free of charge for academic use for installation on Linux or Mac computers.
Questions? Visit our Consed forum.
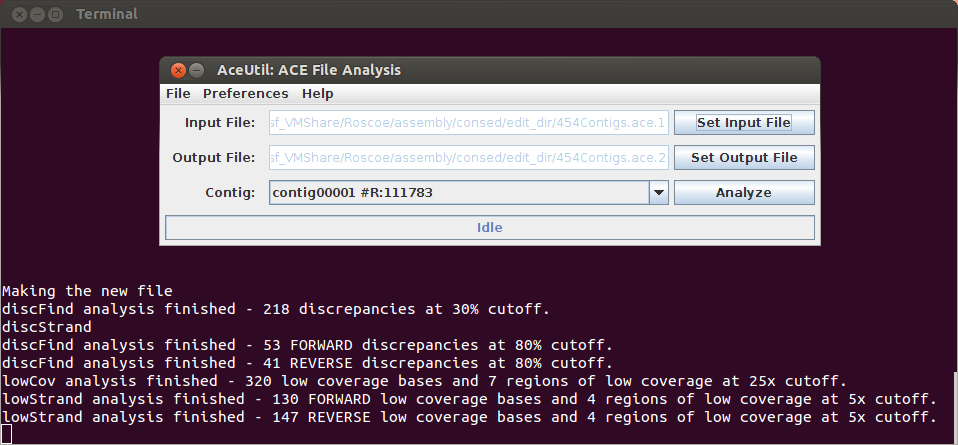
AceUtil is a companion program to Consed. It allows the user to flag consensus positions in a consed assembly based on the percentage of discrepant reads present, or the level of coverage. This streamlines the process of locating potential assembly errors.
AceUtil is written in Java and thus can run on most platforms. It is available as part of the SEA Virtual Machine.
Questions? Visit our AceUtil forum.
Starterator is a program that lets students use comparative genomics data when analyzing auto-annotated gene start sites. Given a gene, Starterator creates a report showing where start site calls have been made for similar genes in other genomes.
Starterator is available as part of the SEA Virtual Machine.
Questions? Visit our Starterator forum.
