Welcome to the forums at seaphages.org. Please feel free to ask any questions related to the SEA-PHAGES program. Any logged-in user may post new topics and reply to existing topics. If you'd like to see a new forum created, please contact us using our form or email us at info@seaphages.org.
Recent Activity
Is this a sequencing error?
| Link to this post | posted 14 Jul, 2021 19:40 | |
|---|---|
|
|
We are annotating genes 8 and 9 in Looper (A15). These two genes are currently orphams. When we look at the same region in similar phages, it's highly conserved and called as one gene (Pham 69334 - 723 members of the pham). When the region from Looper is BLASTed, the alignment shows one nucleotide difference compared to similar phages (see attached), and this results in a stop codon - therefore 2 genes in Looper and one gene in the other phages. There is little coding potential for the second gene in Looper (both programs call it a gene), but the coding potential is minimal for the second half of the gene in similar phages. So the question is, is this a sequencing error? It seems unlikely that one phage out of hundreds would have a mutation leading to a stop codon resulting in two functional genes. |
| Link to this post | posted 15 Jul, 2021 14:25 | |
|---|---|
|
|
Hey Evan, You make a good case as to why this one merits a check as a potential sequencing error, and it has some of those red flags (different from similar genomes, breaks a gene). But I just checked the sequencing data and see this: 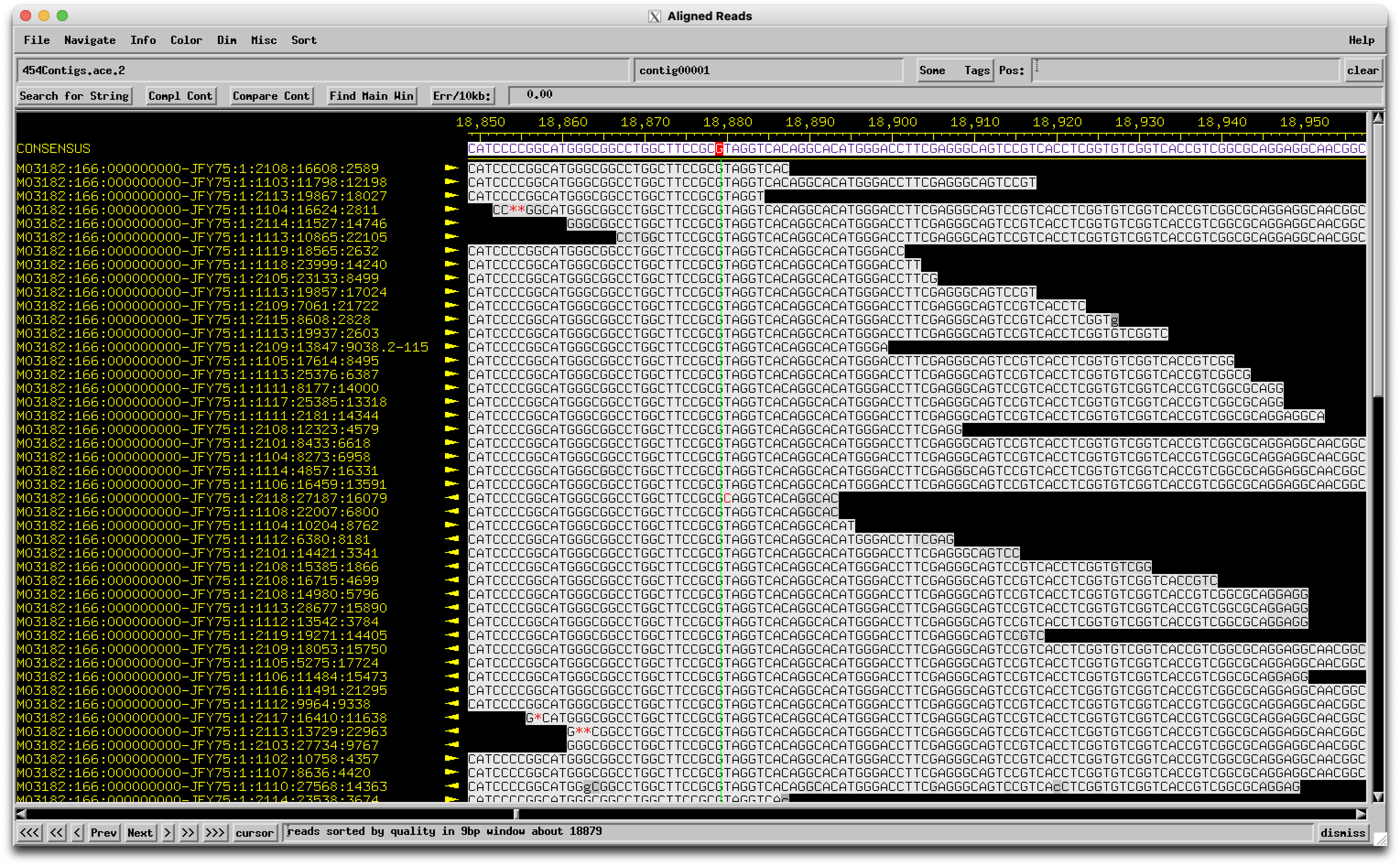 The base in question is a couple to the right of the green line, and the "A" called there is really strongly supported with no conflicting reads. So it's a real biological thing! –Dan |
| Link to this post | posted 15 Jul, 2021 14:32 | |
|---|---|
|
|
DanRussell Great, thanks so much for checking this Dan! |

 78Kb
78Kb