Welcome to the forums at seaphages.org. Please feel free to ask any questions related to the SEA-PHAGES program. Any logged-in user may post new topics and reply to existing topics. If you'd like to see a new forum created, please contact us using our form or email us at info@seaphages.org.
Recent Activity
Phamerator and DNA Master gene numbers don’t match
| Link to this post | posted 02 Mar, 2016 21:15 | |
|---|---|
|
|
Dan, My class recently started annotating Watson and we noticed that the gp numbers on our Phamerator map do not match the auto-annotations in DNA Master (they are off by several numbers after gp8 ). The other genomes we've worked on matched in both programs (until we made changes in DNA Master)and we were wondering if you could explain the difference in the numbering systems? Thank you! Virginia Tech Phage Hunters |
| Link to this post | posted 07 Mar, 2016 15:09 | |
|---|---|
|
|
Hi Stephanie, Sure! First let me say that it’s always possible that any two auto-annotations on the same exact sequence won’t match 100%. This is because of the way the algorithms work and databases change, so if you had 25 students each run an auto-annotation they might not all match. But…the more likely explanation in your case is that it appears that Watson potentially has 3 tRNAs right after gene 8. Since Phamerator doesn’t show tRNAs, you just see a small gap there in Phamerator, though the Phamerator numbering does include those, so it looks like it skips from gene 8 to gene 12. DNA Master doesn’t number the tRNAs by default, so the first gene after them ends up being #9 (which was #12 in Phamerator). You can kind of see the Phamerator and DNA Master maps lined up below. 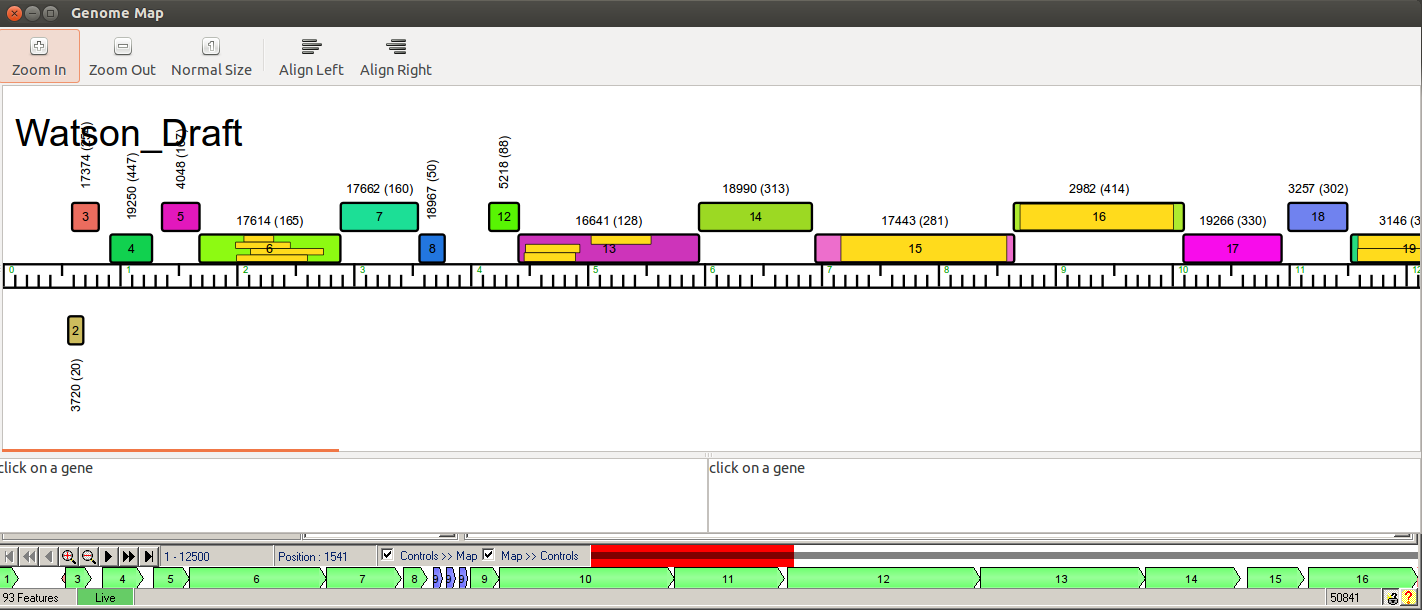 Because of this, we never really work by gene number. We instead use start/stop coordinates to identify genes, because those are unambiguous. And if you add or delete a gene, the gene numbers will eventually change anyway to accommodate the new/deleted genes, so they’re ephemeral until the very final submission. Hope that helps! —Dan |
| Link to this post | posted 09 Mar, 2016 17:54 | |
|---|---|
|
|
Dan, Thank you so much, that makes perfect sense! This situation will help me illustrate the importance of using the start/stop coordinates instead of the gene numbers to my students! -Stephanie |

