Below is a summary of the abstract you submitted. Presenting author(s) is shown in bold.
If any changes need to be made, you can modify the abstract or change the authors.
You can also download a .docx version of this abstract.
If there are any problems, please email Dan at dar78@pitt.edu and he'll take care of them!
This abstract was last modified on April 21, 2015 at 2:32 p.m..

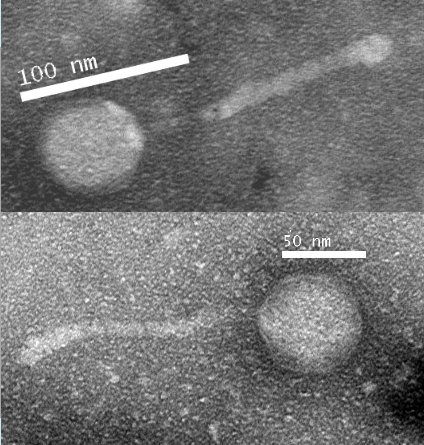
Bacteriophages are viruses that survive by infecting and then replicating using a bacterial host’s genetic machinery, leading to the destruction of the host cell. Bacteriophages’ diversity of attributes, while still being specialized to their host, appeals to the need for continuing research into their survival mechanisms. Bacteriophages and their host bacteria are locked in an evolutionary struggle that has led to the development of new defense mechanisms and new methods of infection. Given the rise in antibiotic resistant bacterial species, bacteriophages may offer a new alternative for combating bacterial infections. During this project, the novel bacteriophage ‘Chupacabra’ was isolated and its genome was sequenced. ’Chupacabra’ belongs to the cluster A and subcluster A10 of bacteriophages and infects Mycobacterium smegmatis. After culturing, ‘Chupacabra’ was revealed to have a temperate life cycle. Each plaque was approximately 3mm in diameter. Classification was assisted by TEM images taken, as well as bioinformatic analysis of phage genomic DNA. The capsid and tail of ‘Chupacabra’ were observed to be 60nm in diameter and 140nm long, respectively. The quality and quantity of DNA harvested from ‘Chupacabra’ were measured through restriction enzyme digest. After DNA sequencing, the ‘Chupacabra’ genome was determined to be 50,286 base pairs in length. Bioinformatic annotation of the ‘Chupacabra’ genome revealed that genes 4 and 46 were unique when compared to other lytic cluster A10 bacteriophages. Hypothesis: The temperate life cycle of ‘Chupacabra’ may be due to differences between ‘Chupacabra’ and other A10 bacteriophages in key DNA synthesis genes.