Welcome to the forums at seaphages.org. Please feel free to ask any questions related to the SEA-PHAGES program. Any logged-in user may post new topics and reply to existing topics. If you'd like to see a new forum created, please contact us using our form or email us at info@seaphages.org.
Recent Activity
PhamDB: Make your own Phamerator databases
| Link to this post | posted 07 Mar, 2016 15:23 | |
|---|---|
|
|
Hi all, Ever wanted to just make your own Phamerator database, but then looked at all the necessary code and immediately given up? It's not an easy process, for sure, but a student named James Lamine from Calvin College—under the supervision of Randy DeJong—has made it much easier. Some of you may remember Randy's talk at last year's Symposium about PhamDB, a web-based way to make your own Phamerator databases. It takes GenBank (.gb) files as input, and outputs links that you can point your copy of Phamerator to in order to use the new database you've created. PhamDB has now been published in Bioinformatics. The installation instructions to get it set up and running on your machine are not bad at all. But we've also set up a (sort of) public version of PhamDB if you want to give it a try. Because it doesn't have account-specific permissions yet, we didn't want to make it fully public, lest random people delete your new databases. So if you're interested in using it, just email me (dar78@pitt.edu) and I'll send you the relevant info. Here's a screenshot: 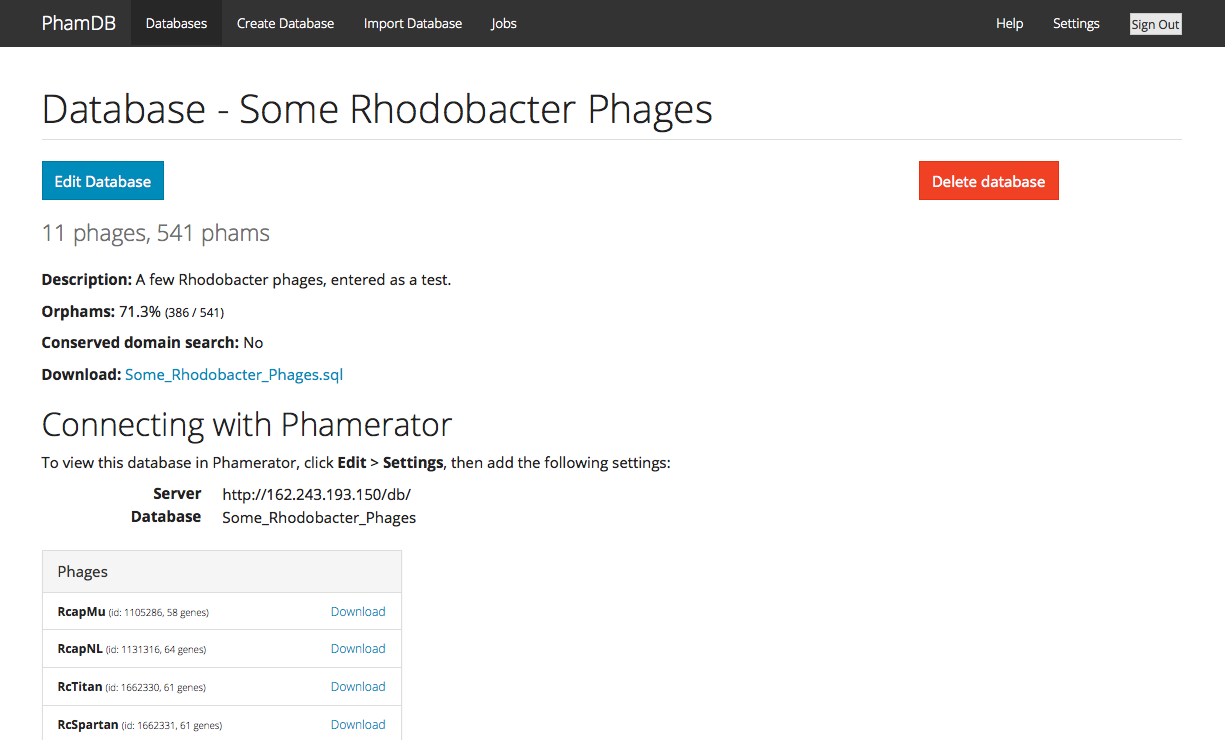 –Dan |
| Link to this post | posted 07 Mar, 2016 16:47 | |
|---|---|
|
|
Just an FYI, you can create the genbank files you need by using the DNA Master "Submit to GenBank" tool which, not surprisingly, is found in the Tools menu. |
| Link to this post | posted 08 Mar, 2016 20:43 | |
|---|---|
|
|
Thanks to Randy and his team for creating the tool and to Dan and Pitt for making a web-based version available. Had great success yesterday with setting up a database for my Streptomyces phages and was able to use the data to create a Splitstree output. This is something that's been on my to-do list for a while, so very happy to be able to do it now! Lee |
| Link to this post | posted 23 Dec, 2016 00:31 | |
|---|---|
|
|
Quick question about PhamDB databases. I have successfully installed it on my machine now (rather than using Dan's web-access version). However, all the phages show up as "singleton" in my database. I am presuming that the Genbank files don't include cluster information, so that is why there is no cluster information. Anyone know how I could edit the GB files so that they include the appropriate cluster information when I load them into PhamDB to generate a Phamerator database? Thanks, Lee |
| Link to this post | posted 03 Feb, 2017 03:52 | |
|---|---|
|
|
Bump - Checking to see if anyone can answer my question about getting cluster information into a PhamDB database (see previous post). Thanks, Lee |
| Link to this post | posted 03 Feb, 2017 17:21 | |
|---|---|
|
|
This was one of the issues that PhamDB did not implement as far as I know. The only way I know to add cluster info is manually (i.e. command line). You can either use a python script that is part of the phamerator distribution or just directly type in mysql commands to update the cluster. There are a lot of specific issues that will depend on your exact computer set-up so impossible to give exact commands. Are you using a SEA virtual machine? |
| Link to this post | posted 03 Feb, 2017 17:33 | |
|---|---|
|
|
I think Chris is right here, and you unfortunately can't add Cluster info via GenBank file, which is the only input for PhamDB. So you're left with the option of locally modifying your database and adding in the clusters. The problem there is that if you then update the database using PhamDB, and your Phamerator imports the newly-updated version, you'll probably lose those changes and have to add them again. Not ideal! –Dan |
| Link to this post | posted 03 Feb, 2017 17:55 | |
|---|---|
|
|
If you are just creating a database that will not be updated then I would recommend just learning a bit of mysql. You might want to check out the mysql help sheet we created at the faculty retreat: https://docs.google.com/document/d/1SUUTojfDKwXCejGY1k14aY92hCqhe8XbDd_-qQPt_gQ/edit?usp=sharing and then just manually update the clusters using command like this: mysql> update phage set Cluster=”S” where Name=”Marvin”; just replace the bold parts with the cluster name and phage name in your database. If you think you will be constantly updating the phamerator database with phamdb I would focus on learning a bit of command line to run the update_clusters.py script (found in the plugins folder of phamerator) which needs a file with phage names and clusters. You can run the script each time after an update with phamdb. |
| Link to this post | posted 03 Feb, 2017 20:41 | |
|---|---|
|
|
Thanks for the information. I'll think about what I need and determine the best solution to use. Lee |
| Link to this post | posted 06 Feb, 2017 16:23 | |
|---|---|
|
|
Hi Lee, on a bit of an unrelated note, would you be willing to share your Streptomyces griseus phage hunting protocols with me? I am getting ready to order this host from ATCC. If you have any tips for success that you are willing to share I would be grateful. I have a group of students doing honors projects that I was going to have do some phage hunting in Great Smoky Mountains National park, where we have had 0 success with m. smeg. |


